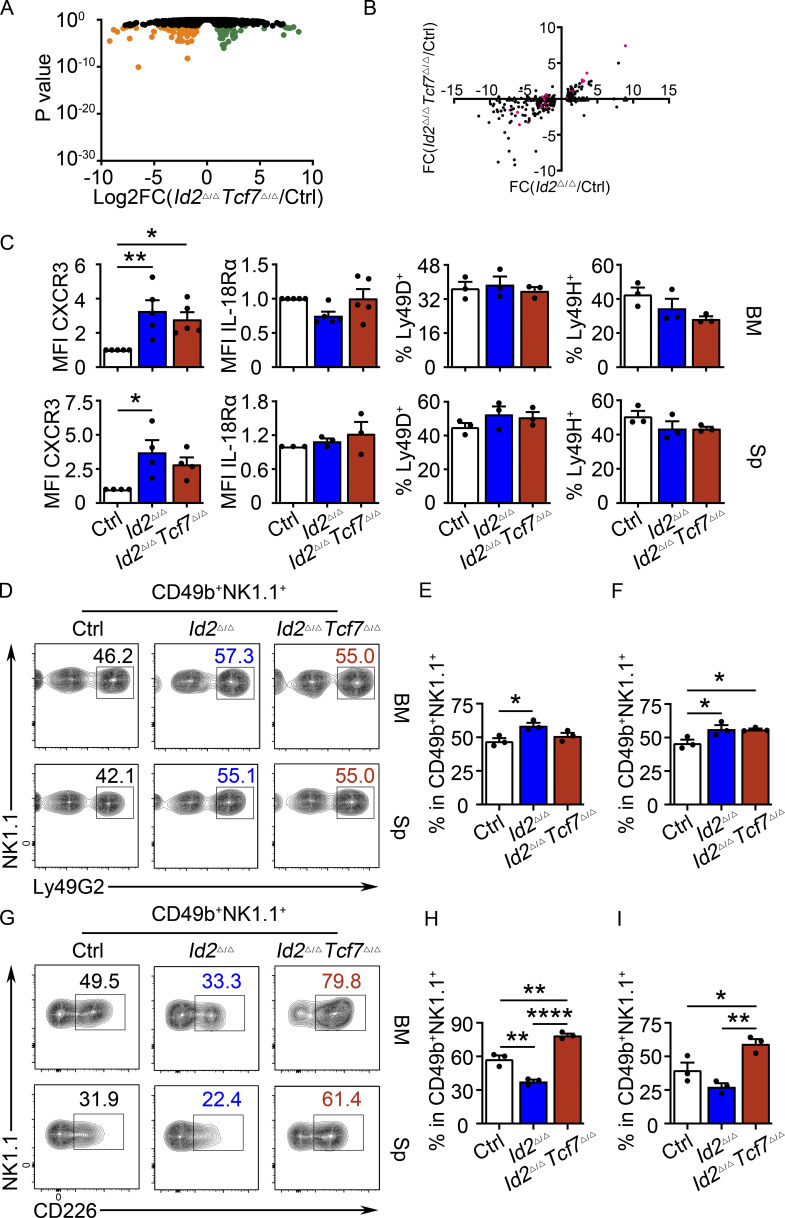
Figure S5.
Partial rescue of NK cell gene expression and surface receptor expression in Id2Δ/ΔTcf7Δ/Δ NK cells. (A) Volcano plot showing Log2FC versus adj. P values for RNA-seq data from Id2Δ/ΔTcf7Δ/Δ and Ctrl NK cells. Data represent three biological replicates. (B) Log2FC for Id2Δ/Δ/Ctrl versus Id2Δ/Δ/Id2Δ/ΔTcf7Δ/Δ for genes that were significant in Id2Δ/Δ/Ctrl (adj. P < 0.05). Each gene is represented by a circle. Pink indicates significance in both comparisons; black indicates significance only in Id2Δ/Δ/Ctrl. (C) Summary of expression of CXCR3, IL-18Rα, Ly49D, and Ly49H on NK cells from BM and spleen (Sp) determined by flow cytometry (n = 3–5 from independent experiments). (D and G) Flow cytometry plots of Ly49G2 and CD226 on NK cells of BM (top) and spleen (bottom) from Id2Δ/ΔTcf7Δ/Δ, Id2Δ/Δ, and Ctrl mice. Numbers are the percentage of cells in the indicated gates (n = 3 from independent experiments). (E and F) Summary of data for Ly49G2 on BM and spleen NK cells. (H and I) Summary of the data for CD226 on BM and spleen NK cells. Error bars represent SEM. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; **, P < 0.01; ****, P < 0.001.