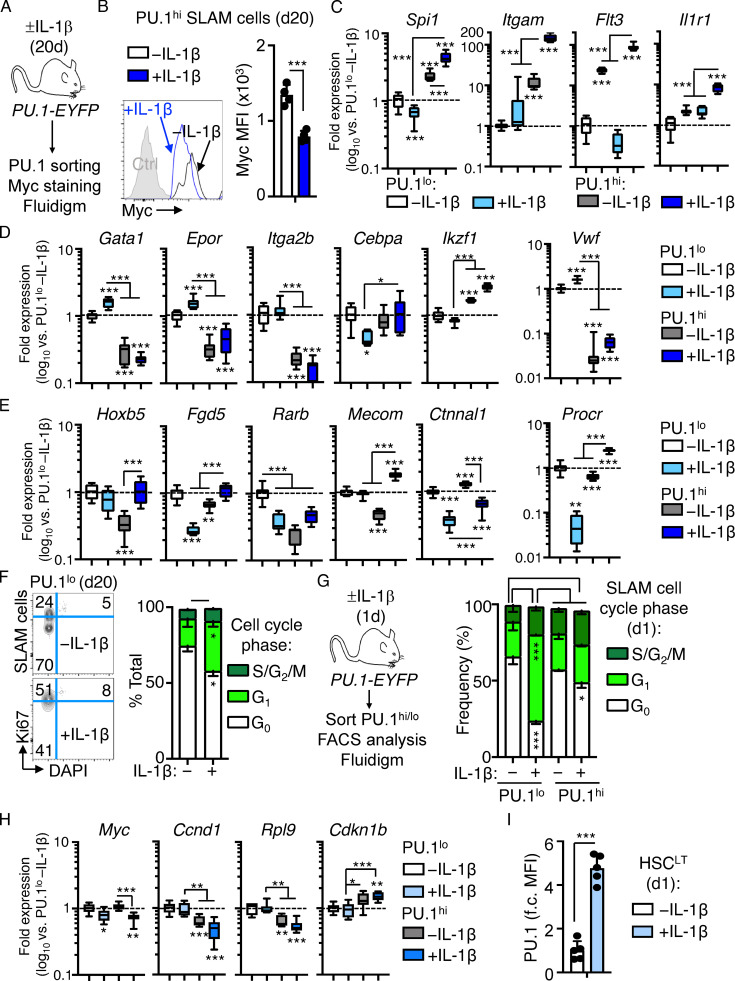
Figure S2.
Characterization of SLAM cell fractions based on PU.1 level. (A) Experimental design for analysis of Myc levels in PU.1hi SLAM cells. (B) Representative FACS plot (left) and quantification (right) of Myc levels in PU.1hi SLAM HSCs from PU.1-EYFP mice treated with or without IL-1β for 20 d (n = 4/group). Individual values are shown with bars representing means. Data are compiled from two independent experiments. (C–E) Quantification by Fluidigm qRT-PCR array of IL-1 target gene expression (C), lineage gene expression (D), and HSC gene expression (E) in PU.1lo and PU.1hi SLAM cells from PU.1-EYFP mice treated with or without IL-1β for 20 d (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (F) Representative FACS plots (left) and quantification (right) of cell cycle distribution in PU.1lo SLAM cells from mice treated with or without IL-1β for 20 d (n = 3/group) using Ki-67 and DAPI. Data are compiled from two independent experiments. (G) Experimental design (left) and quantification (right) of cell cycle distribution in PU.1lo and PU.1hi SLAM cells from mice treated with or without IL-1β for 1 d (n = 4/group). Data are compiled from two independent experiments. (H) Quantification by Fluidigm qRT-PCR array of cell cycle and protein synthesis genes in PU.1lo and PU.1hi SLAM cells from PU.1-EYFP mice treated with or without IL-1β for 1 d (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (I) Quantification of fold change (f.c.) in geometric MFI of HSCLT from PU.1-EYFP mice treated with or without IL-1β for 1 d (n = 5/group). Individual values are shown with bars representing mean values. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Mann-Whitney U test or ANOVA with Tukey’s test in C–E, and H. Error bars represent SD.