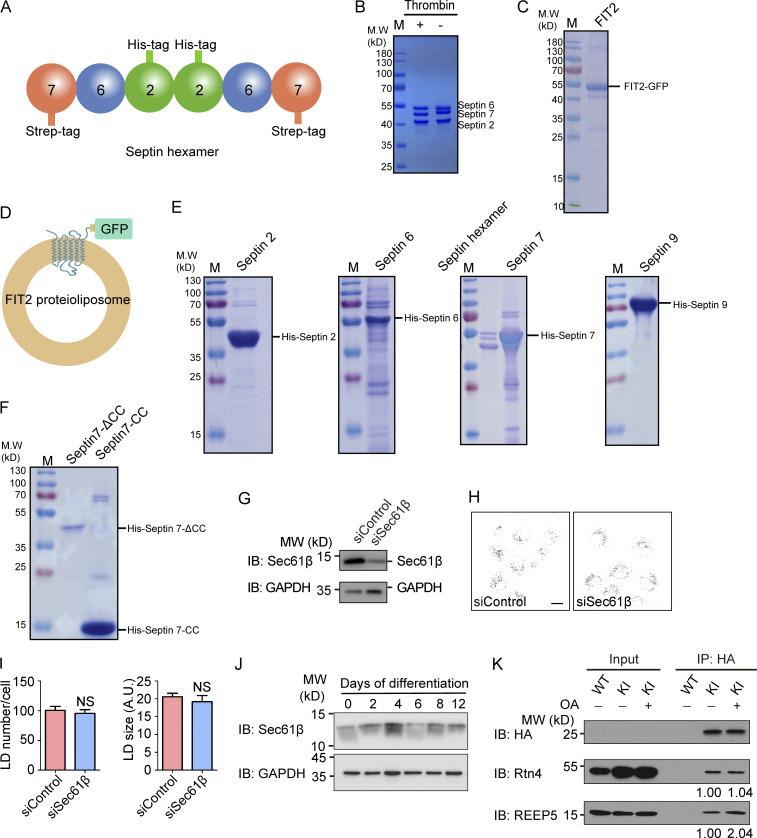
Figure S3.
Purification of septins and FIT2. (A) Representation of the septin2/6/7 hexamer. (B) Purified septin2/6/7 hexamer was detected by SDS-PAGE and Coomassie blue staining. The strep tag on the C-terminus of septin 7 was cleaved by thrombin. (C) The purified FIT2-strep-His8-GFP fusion protein was detected by SDS-PAGE and Coomassie blue staining. (D) Representation of FIT2 proteoliposomes. (E) Purified His-tagged septins 2, 6, 7, and 9 were detected by SDS-PAGE and Coomassie blue staining. (F) Purified His-tagged septin 7 CC and septin 7ΔCC were detected by SDS-PAGE and Coomassie blue staining. (G–I) As in Fig. 5 A but with COS-7 cells depleted of Sec61β. n = 51–88 cells/group. Mann–Whitney test; NS, P > 0.05. Scale bar, 10 µm. Error bars represent SEM. (J) As in Fig. 6 D, Sec61β and GAPDH were detected by immunoblotting (IB). (K) coIP of FIT2 and Rtn4/REEP5 before and after OA treatment. WT and FIT2-HA KI HepG2 cells were delipidated by starving for 60 h or treating with OA for 15 min and lysed in 1% digitonin buffer. IP was performed with anti-HA antibodies, and samples were analyzed by immunoblotting. The relative amount of Rtn4 (coIP) or REEP5 (coIP) compared with FIT2-HA (IP) was quantified by Gel-Pro analyzer software. siSec61β, small interfering Sec61β.