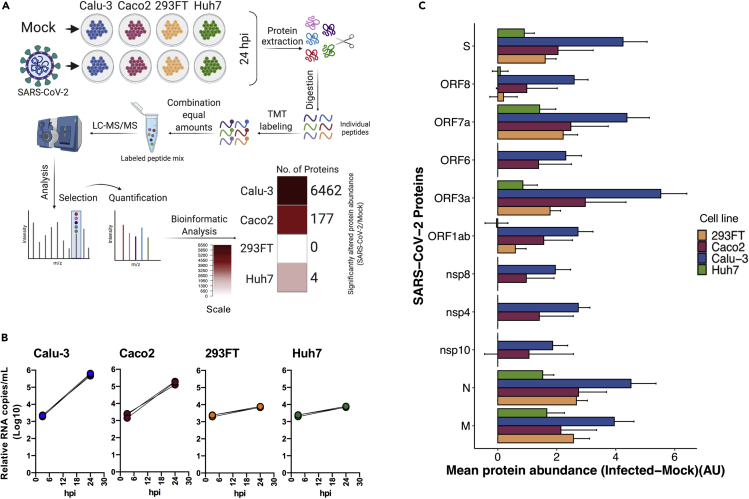
Figure 3.
The proteomic landscape of different commonly used laboratory cell lines upon SARS-CoV-2 infection
(A) Brief methodology of the proteomics experiment in the indicated cell lines that were either mock-infected or infected with SARS-CoV-2 moi 1 for 24 h. The graphical representation was created using BioRender. Significantly altered host proteins in the indicated cell lines are represented in the heatmap.
(B) Relative viral production in the cell supernatant, measured at 3hpi and 24hpi.
(C) Detected viral proteins in the indicated cells by tandem mass tag-labeling mass spectrometry (TMT-MS). The bars represent the difference in viral proteins between infected and uninfected cells (mean abundance of three replicates ± pooled SD) in arbitrary unit (AU). Data were prior quantile normalized.