Abstract
Extra-long chains (ELC) of amylopectin in rice endosperm are synthesized by granule-bound starch synthase I encoded by the Waxy (Wx) gene, which primarily synthesizes amylose. Previous studies showed that single nucleotide polymorphisms (SNP) in intron 1 and exon 6 of the Wx gene influences ELC amount. However, whether these SNPs are conserved among rice cultivars and if any other SNPs are present in the Wx gene remained unknown. Here, we sequenced the Wx gene from 17 rice cultivars with S or L-type amylopectin, including those with known ELC content and those originating in China with unique starch properties, as well as typical japonica and indica cultivars. In addition to the two SNPs described above, an additional SNP correlating with ELC content was found in exon 10. Low ELC cultivars (<3.0 %) had thymine at the splicing donor site of intron 1, Tyr224 in exon 6, and Pro415 in exon 10. Cultivars with moderate ELC content (4.1–6.9 %) had guanine at the splicing donor site of intron 1, Ser224 in exon 6, and Pro415 in exon 10. Cultivars with high ELC content (7.7–13.9 %) had guanine at the splicing donor site of intron 1, Tyr224 in exon 6, and Ser415 in exon 10. The chain length distribution pattern of amylopectin was correlated with the amounts of SSIIa found in starch granules and gelatinization temperature, but not with ELC content. The combinations of SNPs in the Wx gene found in this study may provide useful information for screening specific cultivars with different ELC content.
Keywords: rice, amylose, amylopectin, extra-long chain, Waxy, granule-bound starch synthase
Abbreviations
AAC, apparent amylose content; AGPase, ADP-glucose pyrophosphorylase; BE, starch branching enzyme; DP, degree of polymerization; DSC, differential scanning calorimetry; ELC, extra-long chain; GBSSI, granule-bound starch synthase I; PCR, polymerase chain reaction ; SNP, single nucleotide polymorphism; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SS, starch synthase; Wx, Waxy
INTRODUCTION
Starch is composed of glucose polymers of highly branched amylopectin and essentially linear amylose.1),2) Amylose content, as well as length and frequency of amylopectin branches, largely affects physicochemical properties and functionality of starch.3),4) These in turn determine the commercial application of grains and thus affect the value of grains. Therefore, understanding the mechanisms that control amylopectin branch structure is important.
Amylopectin is synthesized by finely balanced, synergistic actions of multiple isozymes of starch synthases (SS), starch branching enzymes (BE), and starch debranching enzymes.5) SSIIIa synthesizes long amylopectin chains (degree of polymerization (DP) > 30),6) BEI generates long branches in amorphous lamellae,7) and BEIIb generates short branches (DP 6 and 7) in crystalline lamellae.8) The branches generated by BEIIb are elongated to DP 8—12 by SSI9) and can be further elongated to DP 12—24 by SSIIa.10) The length of these amylopectin chains influences the gelatinization11),12) and retrogradation properties of rice starch. 13),14)Amylopectin enriched with short chains (DP ≤10) is designated as S-type while those enriched with longer branches (DP ≤ 24) are designated as L-type amylopectin.15) The extra-long chain (ELC, also called super-long chain) of amylopectin with DP 300–400, the main target of this study, is synthesized by granule-bound starch synthase I (GBSSI), although GBSSI is primarily involved in synthesis of amylose.16),17),18)
GBSSI is encoded by the Waxy (Wx) gene, and the levels of GBSSI protein govern the amylose content and viscoelasticity of cooked rice.19),20) There are several Wx alleles among rice cultivars, including Wxa, Wxb, Wxin, Wxop (or Wxhp), Wxmq and wx, which are assigned to nucleotide polymorphisms of the Wx gene, each giving different levels of apparent amylose content (AAC).20),21),22),23),24),25),26),27),28),29) Wxa is present in most non-glutinous indica rice, which expresses high levels of GBSSI, possesses high amylose (25–30 % of total starch), and has a less sticky texture when cooked.20),30) In contrast, Wxb is present in most non-glutinous japonica rice, which has a single nucleotide polymorphism (SNP) located at the 5′ end of the splicing junction of the first intron of the Wx gene.31),32),33),34) A substitution from GT in Wxa to TT in Wxb causes improper splicing of the pre-mRNA, thus producing less mature GBSSI mRNA and 10-fold less GBSSI protein, resulting in reduced amylose content (15—20 % of total starch), hence giving a slightly sticky texture to cooked rice.31),32),33),34),35),36) Wxin, Wxop, Wxhp, and Wxmq represent a minor proportion of rice cultivars. Wxin, standing for Wx intermediate, is a derivative of the Wxa allele and has an A/C SNP (224th residue Tyr/Ser) in exon 6, resulting in intermediate levels of GBSSI and amylose content.23)Wxop, standing for Wx opaque (or Wxhp, standing for Wx Haopi), is also a derivative of the Wxaallele and has an A/G SNP (Asp166Gly) in exon 4.24),25) This causes a loss of GBSSI affinity to starch granules in Wxhp25), resulting in opaque seeds with very low amylose content (approximately 10 %).24),25),37) Wxmq, standing for Wx Milky Queen, a derivative of the Wxb, has an additional G/A SNP (Arg158His) in exon 4 and a T/C SNP (Tyr191His) in exon 5, resulting in low amylose.26) Glutinous rice has wx allele with a premature termination codon within its coding sequence, resulting in no GBSSI protein, opaque seeds with amylose-free starch, and cooked rice with elastic texture.27),28) In addition, (CT)n repeats located in the untranslated region of exon 1, as well as C/T SNP (415th residue Pro/Ser) in exon 10, are found in the Wx gene of various non-glutinous rice cultivars.38),39),40)
Among the polymorphisms present in the Wx gene, it was speculated that the 5’ end of the splicing junction of intron 1 (GT/TT) and the A/C SNP (224th residue Tyr/Ser) in exon 6 of the Wx gene likely control ELC content.41) Transgenic wx rice expressing high levels of GBSSI with Tyr224 accumulated high levels of ELC, which suggests a correlation between the amount of ELC and the SNP in Wx gene at intron 1 and the residue at 224.41) Two lines with GT at intron 1 and Tyr224 in exon 6 accumulated high ELC; two lines with GT at intron 1 and Ser224 in exon 6 accumulated low ELC; and one line with TT at intron 1 and Tyr224 in exon 6 accumulated low ELC.41) Although extensive studies have shown a correlation between AAC and physicochemical properties of grains, whether the relationship between the levels of ELC content and combinations of SNPs responsible for ELC content is conserved for other rice cultivars remains unknown.
A total of nineteen non-glutinous rice lines as low (< 2 % of total starch), medium (approximately 5–7 % of total starch), or high (> 13 % of total starch) ELC content rice lines were generated by the ‘Super Rice Project’.35) Amount of ELC positively correlated with the setback of starch measured by Rapid Visco Analyzer,35) suggesting that rice with high ELC content may produce harder and stickier rice gel after cooking and cooling. However, whether these rice cultivars have specific types of Wx alleles or unidentified SNPs was unknown.
Therefore, the objectives of this study were to see whether the above-mentioned SNPs are conserved among rice lines with different levels of amylose and ELC and if any other previously unknown SNPs are present in Wx gene. Seven cultivars with known ELC content,41) two glutinous rice cultivars, three typical japonica cultivars, two typical indica cultivars, and four Chinese rice lines with distinct AAC and amylopectin structures were used in this study to sequence the Wx gene and to compare with ELC and AAC. In addition, chain-length distribution patterns of amylopectin branches and gelatinization temperatures were analyzed. Wx gene is located on chromosome 6 near SSI and SSIIa genes in rice, 42)and SNPs present in SSIIa are known to greatly affect amylopectin structure and gelatinization temperature.10),11),12),13),14) This study provides useful information for selecting and/or breeding new rice cultivars with different levels of amylose and ELC that can be used for a variety of food and industrial applications.
MATERIALS AND METHODS
Plant materials.
Oryza sativa L. cvs. EM21 (waxy rice generated by treating Kinmaze with N-methyl-N-nitrosourea),27) BP011,43) BP005 (Zhefu 802),43) Kinmaze,42) Taichung 65,42),44) Nipponbare,41),44),45) Labelle,41),46) Beniroman,41) Chugoku 134,35) Hoshinishiki,35) Hoshiyutaka,35),41) BP028,38) BP003 (Jiayu 293),43) BP037,38) IR 36,41),47) Yumetoiro,35),41) Guizao 2,35) and Tohoku148 (cross between Basmati 370 and Akihikari)35) were used for this study. BP003, BP005, BP011, BP028, and BP037 were grown in an experimental paddy field of Zhejiang University, China, and the rest of the lines were grown during the summer under natural environmental conditions in an experimental paddy field at the Akita Prefectural University, Japan.
Genomic DNA sequencing of the Wx gene.
Genomic DNA was isolated from young seedlings, as described.6) The oligonucleotide sequences for amplification and sequencing are summarized in Supplemental Table 1 and Supplemental Fig. 1. PCR conditions for amplification of the Wx gene were 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s, and 68 °C for 3 min using the Expand Long Template PCR system (Sigma Aldrich, St. Louis, MO, USA former Roche, Basel, Switzerland) supplemented with 2.5 % dimethyl sulfoxide. PCR products were separated by 0.8 % agarose, and the corresponding band was excised and purified using a QIAquick Gel Extraction Kit (Qiagen, Venlo, Netherlands). Purified DNA was sequenced with listed primers (Supplemental Table 1 and Supplemental Fig. 1) using BigDye Terminator (Thermo Fisher Scientific Inc., Waltham, MA, USA former Applied Biosystems, Foster city, CA, USA) at the Biotechnology Center in Akita Prefectural University.
Table 1.
Polymorphisms in the Waxy with guanine at intron 1, Tyr224, and Ser415 resulting in high GBSSI protein, AAC, and ELC regard less of amylopectin types (S or L-type).
| Line | Intron 1 T/G |
Exon 6 224Tyr/Ser |
Exon10 415 Pro/Ser |
Other polymorphism in exon |
GBSSIa | AAC (%) | ELC (%) | SSIIa TBPb |
Amylo- pectin S/L-type |
DSC Tp (oC) |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No ELC | 1 | EM21 | T | Tyr | Pro | Exon 7 Trp235* | - | 0.5±0.1 | ND | - | S | 58.4±0.4 |
| 2 | BP011 | T | Tyr | Pro | Exon 1 -86 (CT)2 Ins, Exon 2 Ins58* |
- | 0.4±0.2 | 0.1±0.0 | ++ | L | 76.2±0.1e | |
| Low ELC | 3 | BP005 | T | Tyr | Pro | --- | + | 11.5±0.3 | 0.7±0.0 | ++ | L | 75.9±0.1e |
| 4 | Kinmaze | T | Tyr | Pro | --- | ++ | 21.5±1.9c | 2.3±0.1c | + | S | 55.4±0.1f | |
| 5 | Taichung 65 | T | Tyr | Pro | --- | ++ | 22.1±0.8c | 2.8±0.1c | + | S | 57.1±0.1f | |
| 6 | Nipponbare | T | Tyr | Pro | --- | ++ | 21.2±0.3 | 3.0±0.4 | + | S | 63.6±0.1 | |
| Medium ELC |
7 | Labelle | G | Ser | Pro | Exon 1 -94 (CT)4 del | +++ | 24.7±0.1 | 4.1±0.1 | ++ | L | 69.0±0.1 |
| 8 | Beniroman | G | Ser | Pro | Exon 1 -88 (CT) del | +++ | 27.6d | 5.1d | ++ | L | 70.3d | |
| 9 | Chugoku 134 | G | Ser | Pro | Exon 1 -86 (CT)2 Ins Exon 10 Ala380Gly Exon 10 Leu434Phe |
+++ | 30.3d | 5.5d | + | S | 64.7d | |
| 10 | Hoshinishiki | G | Ser | Pro | Exon 1 -86 (CT)2 Ins Exon 10 Ala380Gly |
+++ | 29.1d | 5.8d | + | S | 65.9d | |
| 11 | Hoshiyutaka | G | Ser | Pro | Exon 1 -86 (CT)2 Ins | +++ | 28.1d | 6.9d | + | S | 64.9d | |
| High ELC |
12 | BP028 | G | Tyr | Ser | Exon 1 -102 (CT)7 del | +++ | 24.5±1.3 | 7.7±0.4 | ++ | L | 69.9±0.0 |
| 13 | BP003 | G | Tyr | Ser | Exon 1 -102 (CT)7 del | +++ | 24.9±1.0 | 9.9±0.3 | ++ | L | 71.1±0.0 | |
| 14 | BP037 | G | Tyr | Ser | Exon 1 -102 (CT)7 del | +++ | 25.3±1.0 | 10.4±0.6 | + | S | 59.5±0.1 | |
| 15 | IR36 | G | Tyr | Ser | Exon 1 -102 (CT)7 del | +++ | 27.4±0.9 | 10.4 | ++ | L | 70.1±0.1 | |
| 16 | Yumetoiro | G | Tyr | Ser | Exon 1 -102 (CT)7 del | +++ | 29.4d | 13.6d | - | S | 64.7d | |
| 17 | Guizao 2 | G | Tyr | Ser | Exon 1 -102 (CT)7 del | +++ | 31.9d | 13.9d | - | S | 64.7d | |
aAmount of GBSSI in total protein was summarized from Fig. 2. -, +, ++, and +++ indicate no, very low, low, and high GBSSI expression levels, respectively. bAmount of SSIIa in tightly bound to starch granule fraction (TBP) were summarized from Fig. 2. -, +, and ++ indicate no, low, and high SSIIa levels, respectively. cThe data was as previously reported.41) dThe data was as previously reported.35) eHigher gelatinization temperatures than other cultivars with L-type amylopectin. fThe data was as previously reported.60) * indicates a stop codon. ND, not determined.
Fig. 1. Nucleotide polymorphisms of the Waxy gene.
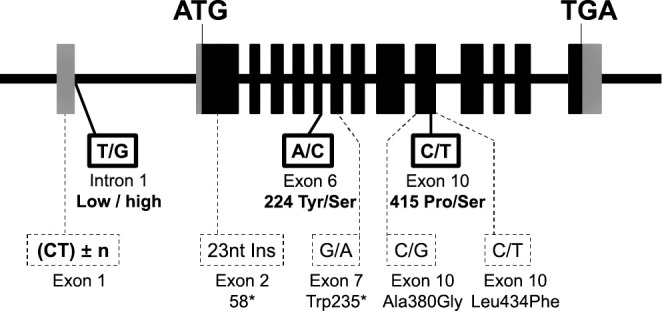
Gray rectangles indicate untranslated regions, and black rectangles indicate coding regions. Common nucleotide polymorphisms that influence extra-long chain content are indicated with black boxes. Other nucleotide polymorphisms within exons are indicated with dotted boxes. * indicates a stop codon.
Purification of starch and amylopectin.
Starches and amylopectin from the powdered mature seeds were purified as described.6),48)
AAC and ELC content.
Gel filtration chromatography of starch and amylopectin was performed as described6),44) using a Toyopearl HW55S gel filtration column (300 × 20 mm, TOSOH, Tokyo, Japan) connected in series to three Toyopearl HW50S columns (300 × 20 mm, TOSOH) and equipped with a refractive index (RI) detector (Tosoh RI-8020, TOSOH). Fractions I, II, and III were separated, and the percentage of Fraction I from total starch was AAC and that of amylopectin was ELC.
Extraction of total proteins, loosely bound, and tightly bound proteins to starch granules, and their western blotting.
The samples were prepared as described previously45) with following modifications. Total proteins were extracted from 10 mg of powdered mature rice seeds using 300 μL of 125 mM Tris–HCl (pH 6.8), 8 M urea, 4 % (w/v) SDS and 5 % (v/v) β-mercaptoethanol, and 0.05 % (w/v) bromophenol blue. Soluble proteins were removed by washing three times with 300 μL of 50 mM imidazole-HCl (pH 7.4), 8 mM MgCl2, 50 mM β-mercaptoethanol, and 12.5 % (v/v) glycerol. Loosely bound proteins to starch granules were extracted three times by 200 μL of 55 mM Tris–HCl (pH 6.8), 10 % SDS, 5 % (v/v) β-mercaptoethanol, and 12.5 % (v/v) glycerol. Tightly bound proteins to starch granules were extracted with 300 μL of 125 mM Tris–HCl (pH 6.8), 8 M urea, 4 % (w/v) SDS, 5 % (v/v) β-mercaptoethanol, and 0.05 % (w/v) bromophenol blue. Loosely-bound to starch granule fractions were pooled, colored with 3 μL of 0.5 % (w/v) bromophenol blue, and denatured by boiling prior to SDS-PAGE. 5 μL of total proteins and tightly bound proteins and 10 μL of loosely bound proteins samples were loaded on 7.5 % acrylamide SDS-PAGE gels. Western blotting was performed using anti-SSI (1:1,000), anti-SSIIa (1:1,000), and anti-GBSSI (1:3,000), as described.49)
Chain-length distribution analyses.
The chain-length distributions of endosperm starch were analyzed by capillary electrophoresis as described50),51) using the P/ACE MDQ Carbohydrate System (AB Sciex, Framingham, MA, USA).
Gelatinization temperature of starch.
Peak gelatinization temperatures were determined by differential scanning calorimetry (DSC6100, Seiko Instruments, Inc., Chiba, Japan) according to the methods described.9)
Prediction of GBSSI protein structure.
Amino acid sequences of GBSSI was deduced from the nucleotide sequences of Guizao 2 and Hoshiyutaka as the representatives of high and medium ELC, respectively. Protein structure was predicted using Phyre2 web portal (http://www.sbg.bio.ic.ac.uk/phyre2)52) and figures were prepared using PyMol software.53)
RESULTS
Nucleotide polymorphisms of the Waxy gene.
To reveal possible nucleotide polymorphisms responsible for ELC content, the Wx gene was sequenced using genomic DNA isolated from seventeen rice lines, including japonica and indica cultivars, eight of which are known to have low, medium, and high AAC and/or ELC as previously reported35),41). Three major single nucleotide polymorphisms were identified and are shown in Fig. 1 as indicated by the bold boxes, two of which resulted in amino acid substitutions. The nucleotide polymorphisms are summarized in Table 1.
The first SNP was the well-known single nucleotide change from guanine to thymine present at the 5′ end of the first intron, where it serves as a splicing donor site. This thymine SNP is known to reduce the expression levels of GBSSI by affecting splicing efficiency.32),33),34) Thymine was present at the 5′ end of the first intron in EM21, BP011, BP005, Kinmaze, Taichung 65, and Nipponbare, while the rest of the lines had guanine.
The second SNP was located in exon 6, and an adenine-to-guanine change led Tyr224 to alter to Ser224. Labelle, Beniroman, Chugoku 134, Hoshinishiki, and Hoshiyutaka had Ser224, while the rest of lines had Tyr224.
The third SNP was located in exon 10, and a cytosine-to-thymine change led Pro415 to alter to Ser415. Ser415 was found in BP028, BP003, BP037, IR36, Yumetoiro, and Guizao 2, and while the rest of lines had Pro415. In addition to these three major SNPs, variations in the number of CT repeats in exon 1 were also found (Table 1), and that deletion of (CT)7 repeats in exon 1 at 102 nucleotides before the start codon was commonly found in BP028, BP003, BP037, IR36, Yumetoiro, and Guizao 2.
Other unconserved nucleotide polymorphisms were as follows: EM21 had a guanine-to-adenine change in exon 7, resulting in Trp235 altering to a stop codon. BP011 had a 23-nucleotide insertion in exon 2, resulting in insertion of a stop codon at the 58th amino acid. Chugoku 134 and Hoshinishiki both had a cytosine-to-guanine change, resulting in Ala380 altering to Gly380; in addition, Chugoku 134 also had an additional cytosine-to-thymine change in exon 10, resulting in Leu434 altering to Phe434. Dozens of unconserved nucleotide polymorphisms were also found in introns, which presumably do not affect expression levels of GBSSI (Data not shown).
Protein expression levels and starch granule affinity of SSI, SSIIa, and GBSSI.
To see whether nucleotide polymorphisms affected the amount of GBSSI, western blotting was performed using total proteins extracted from mature seeds (Fig. 2A). EM21 and BP011 had no GBSSI, since they had early termination codons. Lines with the thymine SNP in intron 1, such as BP005, Kinmaze, Taichung 65, and Nipponbare, had low expression of GBSSI. BP005 had less GBSSI compared to Kinmaze, Taichung 65, and Nipponbare. The lines with the guanine SNP of intron 1 had high expression of GBSSI as expected.
Fig. 2. Protein expression levels and starch granule affinities of SSI, SSIIa, and GBSSI.
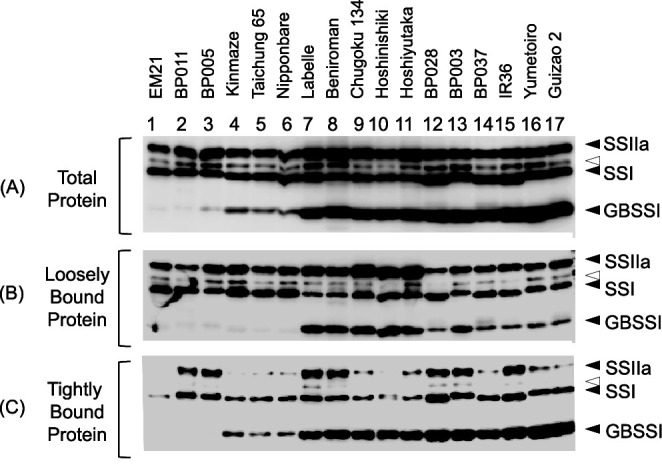
(A) Total protein, (B) protein loosely bound and (C) protein tightly bound to starch granules were extracted from mature seeds. Black arrow heads indicate SSI, SSIIa, and SSIIIa western blot signals, and white arrow heads are non-specific signals. Lanes 1–2, 3–6, 7–11, and 12–17 are rice lines with no, low, medium, and high ELC content, respectively.
Starch granule affinities of SSI, SSIIa, and GBSSI were analyzed by western blotting using protein loosely or tightly bound to the starch granule (Fig. 2B and 2C). Portions of SSIIa in BP011, BP005, Labelle, Beniroman, BP028, BP003, and IR36 were found to be tightly bound to the starch granule (Fig. 2C). SSI was also found to be associated with starch granules, and the proportion of SSI found in tightly bound proteins were greater in BP011, BP005, Labelle, Beniroman, BP028, BP003, and IR36, which was correlated with the association of SSIIa with the starch granules (Fig. 2C). Mobility of SSI on SDS-PAGE gel was slower in BP011, BP005, BP003, Yumetoiro, and Guizao 2 compared with the rest of the lines. This is likely due to the 438th residue of SSI in which Guizao 2 has Glu438, while Nipponbare has Lys438.54) The amount of GBSSI tightly bound to the starch granules (Fig. 2C) in BP028, BP003, BP037, IR 36, Yumetoiro, and Guizao 2 were more than Labelle, Beniroman, Chugoku 134, Hoshinishiki, and Hoshiyutaka. Noticeably, some portion of GBSSI was also found in the lightly bound protein fractions of Labelle, Beniroman, Chugoku 134, Hoshinishiki, Hoshiyutaka, and BP003.
Apparent amylose content (AAC) and extra-long chain (ELC) content.
AAC and ELC are summarized in Table 1. EM21 and BP011 essentially had no amylose or ELC due to a lack of GBSSI. BP005 had low amylose (11 %) and ELC (0.7 %) (Table 1). This outcome is likely due to low expression of GBSSI protein (Fig. 2). The DNA sequence of GBSSI in BP005 was the same as the Kinmaze, Taichung 65, and Nipponbare lines, suggesting that the causal gene for reduced expression of GBSSI is likely to be another gene.
Kinmaze, Taichung 65, and Nipponbare, which all possess thymine at the boundary of intron 1, had relatively low AAC and ELC, 21.2–22.1 and 2.3–3.0 %, respectively (Table 1). The lines with guanine at intron 1, namely, Labelle, Beniroman, Chugoku 134, Hoshinishiki, Hoshiyutaka, BP028, BP003, BP037, IR36, Yumetoiro, and Guizao 2 generally had high AAC (24.5–31.9 %) and medium to high ELC (4.1–13.9 %). This outcome was expected, and all of these lines had high levels of GBSSI (Fig. 2). Although a previous study41) showed that AAC (13 %) and ELC (<1 %) of Labelle was low, our data analyzing the seeds grown under different conditions by different gel filtration systems showed that AAC was 24.7 % and ELC was 4.1 % in Labelle. ELC content was higher in the lines with Tyr224 and Ser415, namely, BP028, BP003, BP037, IR36, Yumetoiro, and Guizao 2, compared with lines with Ser224 and Pro415, namely, Labelle, Beniroman, Chugoku 134, Hoshinishiki , and Hoshiyutaka.
Tohoku 148 (Supplemental Fig. 2) was only the exception, which has thymine at the SNP of intron 1 and Tyr224/Pro415 but with relatively high GBSSI expression levels. Tohoku 148 had high AAC (28.7 %)35) and medium ELC content (6.1 %)35). The reason why Tohoku 148 had high expression of GBSSI may be the deletion of CT at 88 nucleotides prior to the start codon in exon 1, which possibly enabled the proper splicing of the first intron.
Taken together, the results suggest that the combinations of SNPs in GBSSI, particularly residues 224th Tyr/Ser and 415th Pro/Ser, are responsible for ELC content under high expression levels of GBSSI.
Branch structure of starch and gelatinization temperature is correlated with the granule association of SSIIa.
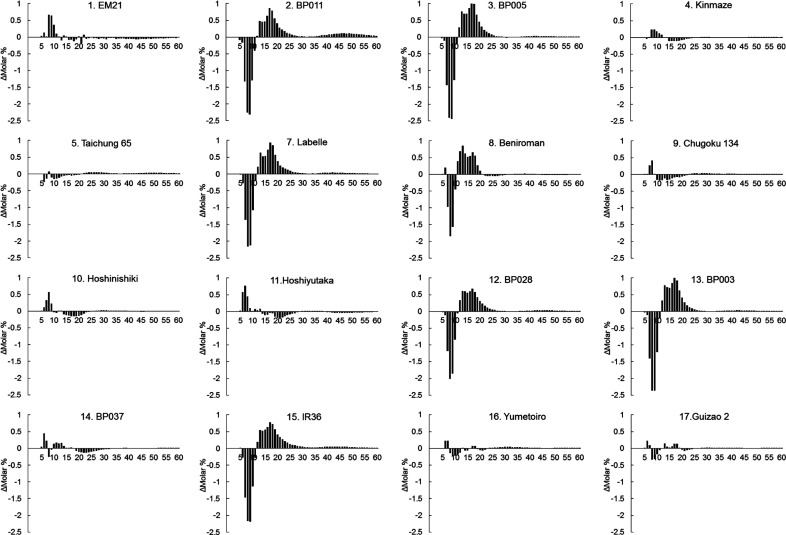
SSI and SSIIa are important genes for determining the structural and physicochemical properties of starch, and they are located near GBSSI on chromosome 6 in rice.42) SNPs present in SSIIa are known to greatly affect its activity and amylopectin structure and hence gelatinization temperature.10),11),12),13),14) Therefore, chain-length distribution of amylopectin was analyzed by capillary electrophoresis using mature seeds. The data was subtracted from that of Nipponbare and shown as a subtraction curve (Fig. 3). Amylopectin structures of EM21, Kinmaze, Taichung 65, Chugoku 134, Hoshinishiki, Hoshiyutaka, BP037, Yumetoiro, and Guizao 2, had only minor differences compared with that of Nipponbare, suggesting these lines had S-type15) amylopectin (Table 1). In contrast, BP011, BP005, Labelle, Beniroman, BP028, BP003, and IR36 had L-type15) amylopectin (Table 1) with fewer short amylopectin chains (DP 6–12) and more medium amylopectin chains (DP 13–24) compared to that of Nipponbare. This peculiar pattern is often associated with differences in activities of SSIIa, where rice with high SSIIa activity has fewer short amylopectin chains and more medium amylopectin chains compared with rice with low SSIIa activity.10),42),55)
Fig. 3. Difference in branch structure of amylopectin compared with Nipponbare having S-type amylopectin.
Chain-length distribution pattern of mature seeds was analyzed by capillary electrophoresis and subtracted from that of Nipponbare. The Y-axis is molar %, and the X-axis is degree of polymerization (DP). Note that BP011, BP005, Labelle, Beniroman, BP028, BP003, and IR36 exhibit branch patterns of L-type amylopectin.
In addition, highly active SSIIa, often seen in typical indica rice, is known to associate with starch granules, while SSIIa of low activity, seen in typical japonica rice, is not found in the tightly bound protein fraction.56),57),58),59) In this study, SSIIa was found to be associated with starch granules in BP011, BP005, Labelle, Beniroman, BP028, BP003, and IR36 (Fig. 2, Table 1).
The gelatinization temperature of rice starch is known to be correlated with the ratio of short and long amylopectin chains governed by SSIIa.11),12),56),57),58),59),60) Therefore, gelatinization temperatures were compared (Table 1). The peak gelatinization temperature of lines with S-type15) amylopectin was low (55-65 °C). In contrast, the peak gelatinization temperature of rice lines with L-type15) amylopectin, namely, BP011, BP005, Labelle, Beniroman, BP028, BP003, and IR36 was high (69-76 °C). These results confirm the relationship between length of amylopectin branches and gelatinization temperature. Among the cultivars with L-type amylopectin, gelatinization temperature of rice cultivars with no or low amylose lines, such as BP011 and BP005, was higher (approximately 76 °C) than the lines with high amylose, such as Labelle, Beniroman, BP028, BP003, and IR36 (approximately 70 °C).
Some of the cultivars analyzed in this study, such as BP005 (SSIIa from indica and Wx from japonica cultivars) and Chugoku 134, Hoshinishiki, Hoshiyutaka, BP037, Yumetoiro, and Guizao 2 (SSIIa from japonica and Wx from indica cultivars), had atypical combinations of SSIIa and Wx genes compared with those from the typical japonica and indica cultivars. This outcome likely resulted from recombination between the SSIIa and Wx genes during a cross between japonica and indica cultivars, which likely lead to accumulation of starch with unique properties.
Comparisons of predicted three-dimensional structure of GBSSI between high and low extra-long chain content.
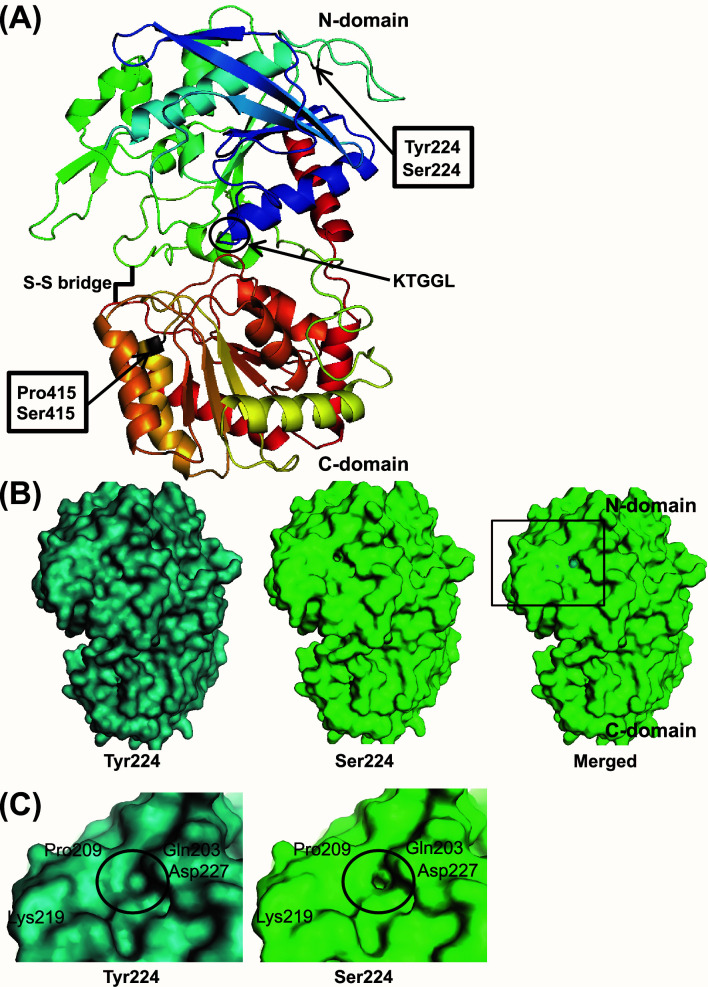
To visualize the effect of polymorphisms in GBSSI on ELC, the predicted three-dimensional structures of GBSSI with Tyr224/Ser415 (high ELC) and Ser224/Pro415 (moderate ELC) were compared. 224th residue Tyr/Ser was located in the N-domain, while 415th residue Pro/Ser was located in at the end of one of the seven alpha helixes present in C-domain closely located to the possible ligand binding site.61) The ribbon models of two lines were completely merged (Fig. 4A), indicating that those polymorphisms at residues 224th Tyr/Ser and 415th Pro/Ser did not affect the backbone structure of GBSSI. However, when the surface models of those two were merged, a difference was observed at 224th residue Tyr/Ser (Fig. 4B and C), but not at 415th residue Pro/Ser. The surface model of the area near Tyr224 in Guizao 2 was filled, while that of Ser224 in Hoshiyutaka was hollow (Fig. 4B and C). Whether this area is involved in association with any ligand is currently unknown.
Fig. 4. Predicted structure of GBSSI by Phyre2.
(A) Ribbon models of GBSSI from high extra-long chain content type (Tyr224/Ser415) and moderate extra-long chain type (Ser224/Pro415) were merged. KTGGL motif and S-S bridge are as shown. (B) Surface model of entire GBSSI. Blue is high extra-long chain content type (Tyr224/Ser415), and green is moderate extra-long chain type (Ser224/Pro415). The area indicated with a black box is zoomed as shown in (C). (C) Zoomed surface model of GBSSI. Note that the black circle indicates the difference near residue 224.
DISCUSSION
The present study using seventeen different rice cultivars with no, low, medium, and high AAC and ELC content levels indicates that expression levels of GBSSI, as well as residue 224 in exon 6 and/or residue 415 in exon 10, are responsible for ELC content in rice (Table 1), partly confirming a previous hypothesis.41) High ELC content cultivars almost always had high levels of GBSSI due to guanine at the SNP of intron 1 and Tyr224/Ser415 (Table 1). High ELC rice cultivars also had deletion of (CT)7 repeats in exon 1 at 102 nucleotides before the start codon. However, this did not affect the expression levels of GBSSI (Fig. 2). (CT)7 repeats may serve as a convenient molecular marker to screen high ELC rice lines. Medium ELC content cultivars had high expression levels of GBSSI due to guanine at the SNP of intron 1 and Ser224/Pro415, but the proportion of GBSSI associated with starch granules were less than that of high ELC content lines (Fig. 2). Low ELC content cultivars had low GBSSI due to thymine at the SNP of intron 1 and Tyr224/Pro415.
Tohoku 148 had thymine at the SNP of intron 1 and Tyr224/Pro415 but with relatively high GBSSI expression levels (Supplemental Fig. 2) resulting in high AAC (28.7 %)35) and medium ELC content (6.1 %).35) The reason why Tohoku 148 had high expression of GBSSI (Supplemental Fig. 2) may be the deletion of CT at 88 nucleotides prior to the start codon in exon 1, which possibly enabled the proper splicing of the first intron.
These outcomes indicate that high GBSSI expression and Tyr224 alone cannot achieve high ELC content and that the combination of Tyr224/Ser415 may also be important. In fact, a mutant lacking SSIIIa isolated from Nipponbare has shown increased expression levels of GBSSI, resulting in high AAC (24.8–30.7 %) and low to moderate ELC (3.1–4.8 %), although rate of increase varied depending on growth condition.6),44) This mutant had an identical Wx gene to Nipponbare with Tyr224/Pro415. This suggests that Ser415 is also an important factor for increasing ELC content.
Previous study analyzing nucleotide polymorphisms of Waxy gene in US and European rice lines showed that AAC and amount GBSSI associated to the starch granules were correlated with combinations of SNPs.62) Guanine at intron 1 and Tyr224/Ser415 showed high AAC while guanine at intron 1 and Ser224/Pro415 showed medium AAC.62) The present study showed that the amount of starch granule associated GBSSI was also correlated with ELC content.
To clarify which amino acids are responsible for high ELC content, one way is to produce and analyze transgenic glutinous rice lines expressing the Waxy gene which has modification in one of these residues.
Although DNA sequence of BP005 was identical to that of Kinmaze, Taichung 65, and Nipponbare, the expression levels of GBSSI were lower, resulting in low AAC and ELC. This shows that expression level of GBSSI controls the amount of ELC. The reason for low GBSSI expression levels in BP005 is yet to be determined. One likely possibility is the presence of a mutation in a transcription factor or splicing factor such as MYC protein,63) ethylene-responsive element-binding protein,63) basic leucine zipper protein,64) splicing factor Ser/Arg-rich proteins,65) and pre-mRNA processing protein.66)
GBSSI is post-translationally regulated, and its activity is likely to be modulated via redox regulation through the formation of S-S bridges and via phosphorylation.67),68),69) Although the effect of post-translational modifications of GBSSI on ELC remains unknown, the involvement of post-translational modifications cannot be excluded, since tyrosine and serine can be phosphorylated. Detailed crystallographic analyses of GBSSI with mutations at amino acid residues 224 and 415, as well as identification of carbohydrate binding modules, may provide further insight into the mechanisms of ELC biosynthesis.
Desired physicochemical properties of starch, such as viscoelasticity and gelatinization temperature, vary depending on the products. The gelatinization temperatures of cultivars with L-type15) amylopectin was generally high (69.0–76.2 °C) compared with that of S-type15) amylopectin (55.4–65.9 °C). However, among the lines with L-type15) amylopectin, the gelatinization temperature was higher for cultivars with no (76.2 °C, BP011) or low AAC (75.9 °C, BP005) than with high amylose (69.0–71.1 °C; IR36, Kasalath, BP003, BP028, Beniroman, and Labelle), confirming the previous study.70) One of the reasons for higher gelatinization temperatures in lines with L-type15) amylopectin with low AAC may be that they can form more uniform double helices in the absence of amylose and ELC. L-type amylopectin with low AAC starch may therefore require higher temperatures to dissociate the helices. Alternatively, GBSSI may also be involved in synthesis of intermediate chains of amylopectin,71) although chain-length distribution patterns of BP005 and BP011 were similar to other L-type amylopectin lines (IR36, Kasalath, BP003, BP028, Beniroman, and Labelle). When amylopectin is bound by GBSSI, amylopectin branches may form slightly looser double helices compared to amylopectin in the absence of GBSSI. Hence, the L-type amylopectin branches bound by high levels of GBSSI may dissociate at lower temperatures. The other possibility is that GBSSI may affect the function of multimeric starch biosynthetic complexes involved in amylopectin biosynthesis, although whether GBSSI regulates the function of the multimeric protein complex is currently unknown.72)
Although the effects of ELC on the viscoelasticity of starch may be minor, high ELC cultivars give high setbacks of starch gel.35) The combinations of nucleotide polymorphisms found in this study may provide useful information for screening specific cultivars with different ELC contents.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the following institutes for kindly providing rice seeds: Furukawa Agricultural Research Station in Miyagi Prefecture for Tohoku 148; NARO Genebank Project for Guizao 2 and Labelle; Kinki Chugoku Shikoku Agricultural Research Center, NARO for Chugoku 134. We also thank the Biotechnology Center in Akita Prefectural University for DNA sequencing. This work was partly supported by the Japan Society for the Promotion of Science [Grant-in-Aid for Young Scientists (B) (#16K18571 to N.C.); Grant-in-Aid for JSPS Research Fellows (#15J40176 to N.C.); Grant-in-Aid for Scientific Research (B) (#19380007 to N.F.)], and President’s Fund of Akita Prefectural University (to N.F.).
REFERENCES
- 1).Hizukuri S.: Polymodal distribution of the chain lengths of amylopectins, and its significance. Carbohydr. Res., 147, 342–347 (1986). [Google Scholar]
- 2).Bertoft E., Koch K., and Man P.: Structure of building blocks in amylopectins. Carbohydr. Res., 361, 105–113 (2013). [DOI] [PubMed] [Google Scholar]
- 3).Jane J., Chen Y.Y., Lee L.F., Mcpherson A.E., Wong K.S., Radosavljevic M., and Kasemsuwan T.: Effects of amylopectin branch chain length and amylose content on the gelatinization and pasting properties of starch. Cereal. Chem., 76, 629–637 (1999). [Google Scholar]
- 4).Syahariza Z.A., Sar S., Hasjim J., Tizzotti M.J., and Gilbert R.G.: The importance of amylose and amylopectin fine structures for starch digestibility in cooked rice grains. Food Chem., 136, 742–749 (2013). [DOI] [PubMed] [Google Scholar]
- 5).Fujita N.: Starch biosynthesis in rice endosperm. AGri-Biosci. Monogra., 4, 1–18 (2014). [Google Scholar]
- 6).Fujita N., Yoshida M., Kondo T., Saito K., Utsumi Y., Tokunaga T., Nishi A., Satoh H., Park J.H., Jane J.L., Miyao A., Hirochika H., and Nakamura Y.: Characterization of SSIIIa-Deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol., 144, 2009–2023 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Satoh H., Nishi A., Yamashita K., Takemoto Y., Tanaka Y., Hosaka Y., Sakurai A., Fujita N., and Nakamura Y.: Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol., 133, 1111–1121 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Nishi A., Nakamura Y., Tanaka N., and Satoh H.: Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol., 127, 459–472 (2001). [PMC free article] [PubMed] [Google Scholar]
- 9).Fujita N., Yoshida M., Asakura N., Ohdan T., Miyao A., Hirochika H., and Nakamura Y.: Function and characterization of starch synthase I using mutants in rice. Plant Physiol., 140, 1070–1084 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Nakamura Y., Francisco P.B., Hosaka Y., Sato A., Sawada T., Kubo A., and Fujita N.: Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol., 58, 213–227 (2005). [DOI] [PubMed] [Google Scholar]
- 11).Bao J.S., Corke H., and Sun M.: Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor. Appl. Genet., 113, 1171–1183 (2006) [DOI] [PubMed] [Google Scholar]
- 12).Waters D.L., Henry R.J., Reinke R.F., and Fitzgerald M.A.: Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol. J., 4, 115–122 (2006). [DOI] [PubMed] [Google Scholar]
- 13).Umemoto T., Aoki N., Lin H., Nakamura Y., Inouchi N., Sato Y., Yano M., Hirabayashi H., and Maruyama S.: Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct. Plant Biol., 31, 671–684 (2004). [DOI] [PubMed] [Google Scholar]
- 14).Xu L., Xie J., Kong X., and Bao J.: Analysis of genotypic and environmental effects on rice starch. 2. Thermal and retrogradation properties. J. Agric. Food Chem., 52 , 6017–6022 (2004). [DOI] [PubMed] [Google Scholar]
- 15).Nakamura Y., Sakurai A., Inaba Y., Kimura K., Iwasawa I., and Nagamine T.: The fine structure of amylopectin in endosperm from Asian cultivated rice can be largely classified into two classes. Starch, 54, 117–131 (2002). [Google Scholar]
- 16).Takeda Y., Hizukuri S., and Juliano B.O.: Structures of rice amylopectins with low and high affinity for iodine. Carbohydr. Res., 168, 79–88 (1987). [Google Scholar]
- 17).Hanashiro I., Matsugasako J., Egashira T., and Takeda Y.: Structural characterization of long unit-chains of amylopectin. J. Appl. Glycosci., 52, 233–237 (2005). [Google Scholar]
- 18).Inouchi N., Hibiu H., Li T., Horibata T., Fuwa H., and Itani T.: Structural characterization of long unit-chains of amylopectin. J. Appl. Glycosci., 52, 239–246 (2005). [Google Scholar]
- 19).Denyer K., Johnson P., Zeeman S., Smith A.M.: The control of amylose synthesis. J. Plant Physiol., 158, 479–487 (2001). [Google Scholar]
- 20).Sano Y.: Differential regulation of waxy expression in rice endosperm. Theor. Appl. Genet., 68, 467–473 (1984). [DOI] [PubMed] [Google Scholar]
- 21).Okagaki R.J.: Nucleotide sequence of a long cDNA from the rice waxy gene. Plant Mol. Biol. 19, 513–516 (1992). [DOI] [PubMed] [Google Scholar]
- 22).Sano Y., Katsumata M., and Okuno K.: Genetic studies of speciation in cultivated rice. 5. Inter- and intraspecific differentiation in the waxy gene expression of rice. Euphytica, 35, 1–9 (1986). [Google Scholar]
- 23).Larkin P.D. and Park W.D.: Association of waxy gene single nucleotide polymorphism with starch characteristics in rice. Mol. Breed., 12, 335–339 (2003). [Google Scholar]
- 24).Mikami I., Uwatoko N., Ikeda Y., Yamaguchi J., Hirano H.Y., Suzuki Y., and Sano Y.: Allelic diversification at the wx locus in landraces of Asian rice. Theor. Appl. Genet., 116, 979–989 (2008). [DOI] [PubMed] [Google Scholar]
- 25).Liu L., Ma X., Liu S., Zhu C., Jiang L., Wang Y., Shen Y., Ren Y., Dong H., Chen L., Liu X., Zhao Z., Zhai H., and Wan J.: Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol., 71, 609–626 (2009). [DOI] [PubMed] [Google Scholar]
- 26).Sato H., Suzuki Y., Sakai M., and Imbe T.: Molecular characterization of Wx-mq, a novel mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.) Breed. Sci., 52, 131–135 (2002). [Google Scholar]
- 27).Isshiki M., Yamamoto Y., Satoh H., and Shimamoto K.: Nonsense-mediated decay of mutant waxy mRNA in Rice. Plant Physiol., 125, 1388–1395 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Wanchana S., Toojinda T., Tragoonrung S., and Vanavichit A.: Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci., 165, 1193–1199 (2003). [Google Scholar]
- 29).Yang B., Xu S., Xu L., You H. and Xiang X.: Effects of Wx and its interaction with SSIII-2 on rice eating and cooking qualities. Front. Plant Sci., 9, 456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Kong X., Zhu P., Sui Z., and Bao J.. . Physicochemical properties of starches from diverse rice cultivars varying in apparent amylose content and gelatinisation temperature combinations. Food Chem., 172, 433–440 (2015). [DOI] [PubMed] [Google Scholar]
- 31).Wang Z.Y., Zheng F.Q., Shen G.Z., Gao J.P., Snustad D.P., Li M.G., Zhang J.L., and Hong MM.: The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J., 7, 613–622 (1995). [DOI] [PubMed] [Google Scholar]
- 32).Cai X.L., Wang Z.Y., Xing Y.Y., Zhang J.L., and Hong M.M.: Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J., 14, 459–465 (1998). [DOI] [PubMed] [Google Scholar]
- 33).Isshiki M., Morino K., Nakajima M., Okagaki R.J., Wessler S.R., Izawa T., and Shimamoto K.: A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5' splice site of the first intron. Plant J., 15, 133–138 (1998). [DOI] [PubMed] [Google Scholar]
- 34).Isshiki M., Nakajima M., Satoh H., and Shimamoto K.: dull: rice mutants with tissue-specific effects on the splicing of the waxy pre-mRNA. Plant J., 23, 451–460 (2000). [DOI] [PubMed] [Google Scholar]
- 35).Horibata T., Nakamoto M., Fuwa H., and Inouchi N.: Structural and physicochemical characteristics of endosperm starches of rice cultivars recently bred in Japan. J. Appl. Glycosci., 51, 303-313 (2004). [Google Scholar]
- 36).Chen M.H., Bergman C.J., Pinson S.R.M., and Fjellstrom R.G.: Waxy gene haplotypes: Associations with pasting properties in an international rice germplasm collection. J. Cereal Sci., 48, 781–788 (2008). [Google Scholar]
- 37).Mikami I., Aikawa M., Hirano H.Y., and Sano Y.: Altered tissue-specific expression at the Wx gene of the opaque mutants in rice. Euphytica, 105, 91–97 (1999). [Google Scholar]
- 38).Bao J., Corke H., and Sun M.: Microsatellites in starch-synthesizing genes in relation to starch physicochemical properties in waxy rice (Oryza sativa L.) Theor. Appl. Genet., 105, 898–905 (2002). [DOI] [PubMed] [Google Scholar]
- 39).Hoai T.T.T., Matsusaka H., Toyosawa Y., Suu T.D., Satoh H., and Kumamaru T.: Influence of single-nucleotide polymorphisms in the gene encoding granule-bound starch synthase I on amylose content in Vietnamese rice cultivars. Breed Sci., 64, 142–148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Li K., Bao J., Corke H., and Sun M M.: Association Analysis of Markers Derived from Starch Biosynthesis Related Genes with Starch Physicochemical Properties in the USDA Rice Mini-Core Collection. Front Plant Sci., 8, 424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Hanashiro I., Itoh K., Kuratomi Y., Yamazaki M,, Igarashi T., Matsugasako J., and Takeda Y.: Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice. Plant Cell Physiol., 49, 925–933 (2008). [DOI] [PubMed] [Google Scholar]
- 42).Umemoto T., Yano M., Satoh H., Shomura A., and Nakamura Y.: Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor. Appl. Genet., 104, 1–8 (2002). [DOI] [PubMed] [Google Scholar]
- 43).Jin L., Lu Y., Xiao P., Sun M., Corke H., and Bao J.: Genetic diversity and population structure of a diverse set of rice germplasm for association mapping Theor. Appl. Genet., 121, 475–487 (2010). [DOI] [PubMed] [Google Scholar]
- 44).Asai H., Abe N., Matsushima R., Crofts N., Oitome N.F., Nakamura Y., and Fujita N.: Deficiencies in both starch synthase IIIa and branching enzyme IIb lead to a significant increase in amylose in SSIIa-inactive japonica rice seeds. J. Exp. Bot., 65, 5497–5507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Abe N., Asai H., Yago H., Oitome N.F., Itoh R., Crofts N., Nakamura Y., Fujita N.: Relationships between starch synthase I and branching enzyme isozymes determined using double mutant rice lines. BMC Plant Biol., 14, 80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Bollich C.N., Atkins J.G., Scott J.E., and Webb B.D.: Registration of Labelle Rice1 (Reg. No. 38). Crop Sci., 13, 773–774 (1973). [Google Scholar]
- 47).Ballini E., Berruyer R., Morel J.B., Lebrun M.H., Nottéghem J.L., and Tharreau D.: Modern elite rice varieties of the ‘Green Revolution’ have retained a large introgression from wild rice around the Pi33 rice blast resistance locus. New Phytol., 175, 340–350 (2007) [DOI] [PubMed] [Google Scholar]
- 48).Fujita N., Toyosawa Y., Utsumi Y., Higuchi T., Hanashiro I., Ikegami A., Akuzawa S., Yoshida M., Mori A., Inomata K., Itoh R., Miyao A., Hirochika H., Satoh H., and Nakamura Y.: Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. J. Exp. Bot., 60, 1009–1023 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Crofts N., Abe N., Oitome N.F., Matsushima R., Tetlow I.R., Emes M.J., Nakamura Y., and Fujita N.: Amylopectin biosynthetic enzymes from rice developing seed form enzymatically active protein complexes. J. Exp. Bot., 66, 4469–4482 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).O'Shea M.G. and Morell M.K.: High resolution slab gel electrophoresis of 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis, 17, 681–686 (1996). [DOI] [PubMed] [Google Scholar]
- 51).Fujita N., Hasegawa H., and Taira T.: The isolation and characterization of a waxy mutant of diploid wheat (Triticum monococcum L.). Plant Sci., 160, 595–602 (2001). [DOI] [PubMed] [Google Scholar]
- 52).Kelley L.A., Mezulis S., Yates C.M., Wass M.N., and Sternberg M.J.: The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc., 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).DeLano W.L.: Pymol: An open-source molecular graphics tool. CCP4 Newsletter On Protein Crystallography, 40, 82–92 (2002). [Google Scholar]
- 54).Chen Y. and Bao J.: Underlying mechanisms of zymographic diversity in starch synthase I and pullulanase in rice-developing endosperm. J. Agric. Food Chem., 64, 2030–2037 (2016). [DOI] [PubMed] [Google Scholar]
- 55).Yu G., Olsen K.M., and Schaal B.A.: Association between nonsynonymous mutations of starch synthase IIa and starch quality in rice (Oryza sativa). New Phytol., 189, 593–601 (2011). [DOI] [PubMed] [Google Scholar]
- 56).Umemoto T. and Aoki N.: Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Funct. Plant Biol., 32, 763–768 (2005). [DOI] [PubMed] [Google Scholar]
- 57).Bao J., Xiao P., Hiratsuka M., Sun M., and Umemoto T.: Granule-bound SSIIa protein content and its relationship with amylopectin structure and gelatinization temperature of rice starch. Starch, 61, 431–437 (2009). [Google Scholar]
- 58).Luo J., Ahmed R., Kosar-Hashemi B., Larroque O., Butardo V.M., Tanner G.J., Colgrave M.L., Upadhyaya N.M., Tetlow I.J., Emes M.J., Millar A., Jobling S.A., Morell M.K., and Li Z.: The different effects of starch synthase IIa mutations or variation on endosperm amylose content of barley, wheat and rice are determined by the distribution of starch synthase I and starch branching enzyme IIb between the starch granule and amyloplast stroma. Theor. Appl. Genet., 128, 1407–1419 (2015). [DOI] [PubMed] [Google Scholar]
- 59).Lu Y., Xiao P., Shao Y., Zhang G., Thanyasiriwat T., Bao J.: Development of new markers to genotype the functional SNPs of SSIIa, a gene responsible for gelatinization temperature of rice starch. J. Cereal Sci., 52, 438–443 (2010). [Google Scholar]
- 60).Abe N., Nakamura Y., and Fujita N.: Thermal properties, morphology of starch granules, and crystallinity of endosperm starch in SSI and BE isozyme double mutant lines. J. Appl. Glycosci., 60, 171–176 (2013). [Google Scholar]
- 61).Nielsen M.M., Ruzanski C., Krucewicz K., Striebeck A., Cenci U., Ball S.G., Palcic M.M., and Cuesta-Seijo J.A.: Crystal Structures of the Catalytic Domain of Arabidopsis thaliana Starch Synthase IV, of Granule Bound Starch Synthase from CLg1 and of Granule Bound Starch Synthase I of Cyanophora paradoxa Illustrate Substrate Recognition in Starch Synthases. Front Plant Sci., 9, 1138 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Dobo M., Ayres N., Walker G., and Park W.D.: Polymorphism in the GBSS gene affects amylose content in US and European rice germplasm. J. Cereal Sci., 52, 450–456 (2010) [Google Scholar]
- 63).Zhu Y., Cai X.L., Wang Z.Y. and Hong M.M.: An Interaction between a MYC Protein and an EREBP Protein Is Involved in Transcriptional Regulation of the Rice Wx Gene. J. Biol. Chem., 278, 47803–47811 (2003). [DOI] [PubMed] [Google Scholar]
- 64).Wang J.C., Xu H., Xhu Y., Liu Q.Q., and Cai X.L.: OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot., 64, 3453–3466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Isshiki M., Tsumoto A., and Shimamoto K.: The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell, 18, 146–158 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Zeng D., Yan M., Wang Y., Liu X., Qian Q., and Li J.: Du1, encoding a novel Prp1 protein, regulates starch biosynthesis through affecting the splicing of Wxb pre-mRNAs in rice (Oryza sativa L.). Plant Mol. Biol., 65, 501–509 (2007). [DOI] [PubMed] [Google Scholar]
- 67).Momma M. and Fujimoto Z.: Interdomain disulfide bridge in the rice granule bound starch synthase I catalytic domain as elucidated by X-ray structure analysis. Biosci. Biotechnol. Biochem., 76, 1591–1595 (2012). [DOI] [PubMed] [Google Scholar]
- 68).Liu D.R., Huang W.X., and Cai X.L.: Oligomerization of rice granule-bound starch synthase 1 modulates its activity regulation. Plant Sci., 210, 141–150 (2013). [DOI] [PubMed] [Google Scholar]
- 69).Wang S.J. , Liu L.F. , Chen C.K. , and Chen L.W. : Regulations of granule-bound starch synthase I gene expression in rice leaves by temperature and drought stress. Biol. Plantarum 50, 537–541 (2006). [Google Scholar]
- 70).Xu F., Zhang G., Tong C., Sun X., Corke H., Sun M., and Bao J.: Association mapping of starch physicochemical properties with starch biosynthesizing genes in waxy rice (Oryza sativa L.). J. Agric. Food Chem., 61, 10110–10117 (2013). [DOI] [PubMed] [Google Scholar]
- 71).Zhang C., Chen S., Ren X., Lu Y., Liu D., Cai X., Li Q., Gao J., and Liu Q.: Molecular structure and physicochemical properties of starches from rice with different amylose contents resulting from modification of OsGBSSI activity. J. Agric. Food Chem., 65, 2222–2232 (2017). [DOI] [PubMed] [Google Scholar]
- 72).Bao J.S.: Toward understanding the genetic and molecular bases of the eating and cooking qualities of rice. Cereal Foods World, 57, 148–156 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.