Figure 4. Drug repositioning candidates for COVID-19.
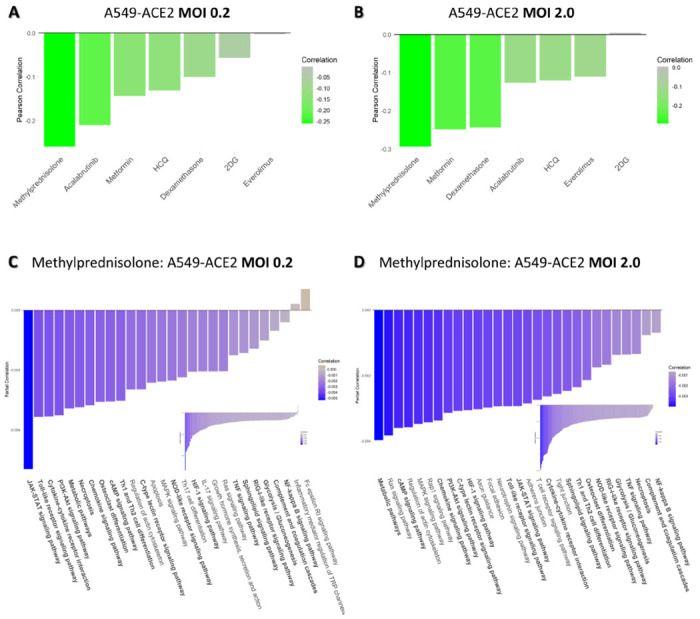
We leverage our PHENSIM drug strategy approach to test candidate drugs for potential repurposing for COVID-19 treatment. Once a cell-specific viral signature is defined, it can be exploited to search for possible repositioning candidates by building a drug signature database. A Pearson correlation ρ(x,y) between the viral and drug signatures gives rise to a correlation score. Drug candidates having a positive effect on ameliorating SARS-CoV-2 infection have a negative correlation score (green) between viral and drug signature, whereas candidate drugs worsening disease correlate positively (red). Here we show distinct candidate drugs having a variable effect depending on the multiplicity of infection (MOI) of virus infection in A459-ACE2 expressing cells in A) low MOI 0.2 and B) high MOI 2.0. This analysis shows the modeling viral load dynamics and discerning what candidate could work best in low vs higher viral load. Resulted top pathways significantly affected by Methylprednisolone treatment are depicted for C) low MOI 0.2 and D) high MOI 2.0. Drug candidates represented here: Methylprednisolone, Metformin (mTOR-inhibitor), (Hydroxy)chloroquine (HCQ-CQ), Acalabrutinib (BTK-inhibitor), Dexamethasone, 2-Deoxy-Glucose (2DG) and Everolimus (mTOR-inhibitor). ACE2; angiotensin-converting enzyme, MOI; multiplicity of infection.
