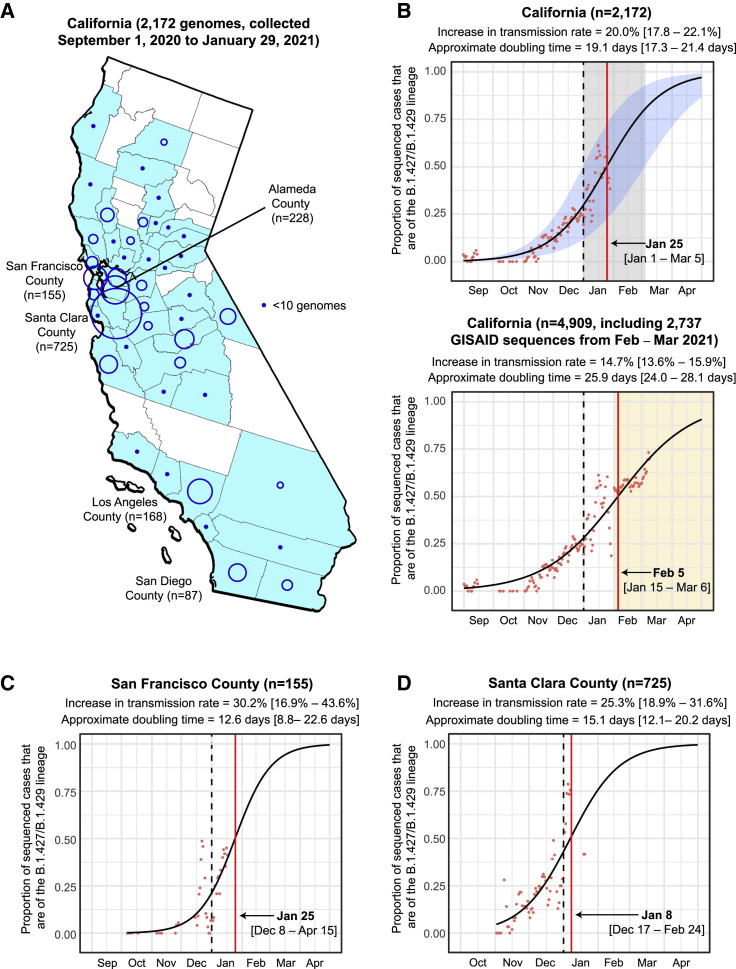
Figure 1.
Increasing frequency of the B.1.427/B.1.429 variant in California from September 1, 2020 to January 29, 2021
(A) County-level representation of the 2,172 newly sequenced SARS-CoV-2 genomes in the current study. Counties from which at least 1 genome were sequenced are colored in sky blue. The size of the circle is proportionate to the number of genomes sequenced from each county, while points designate counties where fewer than 10 genomes were sequenced.
(B–D) Logistic growth curves fitting the 5-day rolling average of the estimated proportion of B.1.427/B.1.429 variant cases in (B) California, (C) San Francisco County, and (D) Santa Clara County. For each curve, the estimated increase in transmission rate and doubling time are shown, along with their associated 95% confidence intervals. The predicted time when the growth curve crosses 0.5 is indicated by a vertical red line. A vertical black dotted line denotes the transition from 2020 to 2021. (B) Top: the logistic growth curve generated from all 2,172 genomes in the current study. The 95% confidence intervals for the increase in transmission rate and doubling time are shaded in blue and gray, respectively. (B) Bottom: the logistic growth curve with inclusion of an additional 2,737 sequenced genomes from California collected February 1 to March 11, 2021. The increase in transmission rate is defined as the logistic growth rate multiplied by the serial interval (Volz et al., 2020; Washington et al., 2021).
See also Figures S1 and S2 and Tables S1, S2, and S5.