Abstract
Background
Long non-coding RNAs (lncRNAs) are implicated in cancer-related biological processes such as cell proliferation, cell cycle progression, cell migration, cell invasion, and chemoresistance. However, the effects of the lncRNA ZEB1-AS1 on oral squamous cell carcinoma (OSCC) have not been adequately demonstrated. The aims of our current study were to explore the roles of lncRNA ZEB1-AS1 in OSCC progression to reveal the potential mechanism.
Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to measure relative ZEB1-AS1 expression levels in OSCC tissues and adjacent non-cancerous tissues. The biological functions of ZEB1-AS1 in OSCC growth and progression were identified by cell proliferation, wound healing, and in vitro transwell assays as well as in vivo xenograft model. The underlying mechanism was detected with a dual-luciferase reporter (DLR) assay.
Results
The up-regulation of ZEB1-AS1 and downregulation of miR-23a-3p (miR-23a) were found in OSCC cancer tissues. A ZEB1-AS1 knockdown remarkably suppressed in vitro cancerous, biological processes of OSCC cell lines such as cell proliferation, invasion, migration, and epithelial–mesenchymal transition (EMT). The tumor growth was also inhibited by silencing ZEB1-AS1 in vivo, and a DLR assay confirmed the association between ZEB1-AS1 and miR-23a.
Conclusion
The newly identified lncRNA ZEB1-AS1 functions as a tumor promoter in OSCC through regulation of miR-23a. Based on these results, ZEB1-AS1 could be a valid molecular target for treating oral cancer.
Keywords: OSCC, lncRNA ZEB1-AS1, miR-23a, EMT
Introduction
Oral squamous cell carcinoma (OSCC) is the sixth most common malignant tumor in the world, and its morbidity and mortality have increased rapidly over the past decades.1 OSCC comprises approximately 3% of all newly diagnosed clinical cancer cases annually.2 Smoking, alcohol or areca abuse, and human papillomavirus (HPV) infections are the leading risk factors for OSCC.3 The rapid progression and invasive growth of OSCC mean that more than 60% of OSCC patients are diagnosed at an advanced stage and receive a poor prognosis.4,5 The 5-year overall survival (OS) rate of patients with OSCC is estimated to be less than 50% though considerable improvements have been made in surgical techniques, chemotherapy, radiotherapy, and immunotherapy.6 Accumulating evidence demonstrates that the main obstacles in OSCC treatment are local recurrence and distant metastasis. Therefore, it is critical to investigate the molecular mechanisms related to OSCC recurrence and metastasis for a more functional cancer therapy approach.
Long non-coding RNAs (lncRNAs) are novel regulators that are more than 200 nucleotides long and have limited protein-coding abilities. Their presence has been demonstrated in multiple biological processes, including metabolism, migration, apoptosis, cell proliferation, and genomic stability.7,8 Recently emerging evidence suggests that aberrant lncRNA expression levels occur frequently in human cancers, and these findings indicate that lncRNAs participate in tumor growth, angiogenesis, and metastasis.9 For instance, Li et al demonstrated that the lncRNAs AC026904.1 and UCA1 contribute to breast cancer metastasis by modulating the TGF-β-induced epithelial-mesenchymal transition (EMT) progress.10 Ni et al reported that the progression of hepatocellular carcinoma (HCC) can be inhibited by lncRNA uc.134 by suppressing LATS1 ubiquitination, which is mediated by CUL4A.11 Therefore, understanding the potential role of lncRNAs in tumors represents a new direction to develop anti-cancer therapeutic strategies.
Recently, the role of ZEB1-AS1 as an oncogene has been determined in human cancers including lung adenocarcinoma, esophageal squamous cell carcinoma (ESCC), and HCC.12–14 The lncRNA ZEB1-AS1 promotes tumor growth and metastasis by accelerating cell proliferation, migration, and tumor angiogenesis.14 Moreover, a study recently revealed that increased ZEB1-AS1expression levels are present in cervical cancer and may be involved in EMT progress.15 However, the exact mechanisms of ZEB1-AS1-mediated OSCC proliferation and progression are still unclear. LncRNAs commonly exert their functions in tumorigenesis and tumor progression by interacting with miRNAs and modulating the expression of miRNA downstream targets. Accumulating evidences indicate that miRNA-23a may be a critical regulator in carcinogenesis and aberrant miR-23a expression has been detected in many cancers.16,17 Advances in cancer research have highlighted the cancer-promoting function of miR-23a in regulating cell proliferation, apoptosis, EMT and angiogenesis progress16,18 However, some recent studies reported that miR-23a is downregulated in certain cancer types, including nephroblastomas and osteosarcoma.19,20 Another group of researchers showed that miR-23a can exhibit pro-apoptotic functions.21 Therefore, the detailed role(s) and molecular mechanism(s) of miR-23a in carcinogenesis still need to be studied.
Our study demonstrates that ZEB1-AS1 expression levels are markedly upregulated in OSCC tissues and correlates with tumor progression. The molecular mechanism of ZEB1-AS1 in OSCC was explored using loss-of-function experiments. Our results showed that OSCC cell proliferation, invasion, and migration in vitro can be suppressed by ZEB1-AS1 knockdown. We also demonstrated that OSCC tumor growth and EMT can be inhibited by silencing ZEB1-AS1 in vivo. Further mechanistic analysis revealed that ZEB1-AS1 is as a ceRNA of miR-23a. Our current study contributes to the potential functions of ZEB1-AS1 in OSCC, which can be considered a novel candidate for future oral cancer treatments.
Materials and Methods
Human Clinical Samples
The China Medical University provided 30 fresh OSCC tissues and adjacent non-cancer tissues collected from June 1, 2019, to December 30, 2019. The enrolled clinical samples met two inclusion criteria: (i) The pathological diagnosis is OSCC. (ii) The patient did not receive anti-cancer therapy (radiotherapy, chemotherapy, or biotherapy) before the surgery. For the 5‐year follow‐up, a total of 88 liquid-nitrogen-preserved tissue specimens from patients with OSCC were provided by our hospital’s biological sample bank. All patients were informed of the complete experimental design and provided written informed consent before enrolling in the study. The tumor node metastasis (TNM) classification system of the Union for International Cancer Control (UICC) was used to determine primary tumor stages and the American Joint Committee on Cancer (AJCC) Criteria were applied to classifying the histological tumor grades of patient samples (Table 1). The median of lncRNA ZEB1-AS1 expression (ΔCt=10.38) was used as a criterion to classify the gene expression level (high or low).
Table 1.
The Clinical Characteristics of the OSCC Patients
| Features | Patient Number | ZEB1-AS1 Expression | P value | |
|---|---|---|---|---|
| n=30 | Low | High | ||
| Age (years) | 0.0253* | |||
| ≥60 | 18 | 6 | 12 | |
| <60 | 12 | 9 | 3 | |
| Gender | 1.0000 | |||
| Female | 15 | 7 | 8 | |
| Male | 15 | 8 | 7 | |
| Tumor size | 0.2723 | |||
| ≥3cm | 16 | 6 | 10 | |
| <3cm | 14 | 9 | 5 | |
| TNM stage | 0.0209* | |||
| I–II | 19 | 13 | 6 | |
| III–IV | 11 | 2 | 9 | |
| Differentiation | 0.0022** | |||
| Poorly | 22 | 7 | 15 | |
| Moderately well | 8 | 8 | 0 | |
Notes: For analysis of correlation between ZEB1-AS1 expression and clinical features, fisher exact tests were used. *P < 0.05; **P < 0.001.
Abbreviations: OSCC, oral squamous cell carcinoma; TNM, tumor node metastasis.
Cell Culture and Transfection
The OSCC cell lines (SCC9, SCC25, TSCCA, HN4, and CAL27) and the human normal oral epithelial cell line HOK were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). Cell cultures were grown at 37°C in Dulbecco’s modified Eagle’s medium/F12 (DMEM/F12; Gibco, MD, USA) with 10% fetal bovine serum (FBS; Gibco, MD, USA) in a humidified atmosphere (5% CO2, 95% air). Short-hairpin RNA (shRNA) specially targeted to ZEB1-AS1 (sh-ZEB1-AS1), control shRNA (sh-NC), as well as mimics, inhibitors, and NC of miR-23a were obtained from Genepharma Corp., Ltd. (Shanghai, China). A 6-well plate with antibiotic-free medium was seeded with cells (5×105 cells/well) in the logarithmic phase before transfection. Lipofectamine 3000 (Invitrogen, CA, USA) was applied to transfect the oligonucleotides (50 pmol/mL) per manufacturer protocols. Experiments were conducted on the transfected cells 48h post-transfection.
Cell Cycle Analysis
Flow cytometry analyses were performed to detect the cell cycle distribution. Briefly, a standard trypsinization procedure was used to collect the transfected cells 48h after shRNA transfection, and cells were fixed with 75% ethanol overnight at 4°C. RNase A work solution (0.01 mg/mL) and propidium iodide (PI) solution (0.05 mg/mL) were then applied and incubated at room temperature (RT) for 30 min in the dark. The stained cells were analyzed by flow cytometry using a FACSCalibur system (BD Biosciences, CA, USA), and MultiCycle software (Phoenix Flow Systems, CA, USA) was used to analysis cell cycle distribution. The experiments in each group were completed in triplicate.
Cell Proliferation Assay
EdU cell proliferation assay was performed using a Click-iT EdU assay kit (Invitrogen, CA, USA) per manufacturer protocols. Briefly, the 6-well plates were seeded with cells (5×104 cells/well) in logarithmic phase and cultured for 24h at 37 °C. Cells were incubated in EdU work solution (20 mM) at RT for another 2h. DAPI (1: 1000, Invitrogen, CA, USA) was used to stain the nuclei of cells fixed with 4% paraformaldehyde (PFA) following EdU incubation. An inverted fluorescent microscope (Leica DMI6000B, Wetzlar, Germany) was used to image EdU positive cells. Independent experiments were completed in triplicate.
Wound-Healing Assay
The migratory abilities of sh-ZEB1-AS1-transfected OSCC cells were determined using a wound-healing assay. Briefly, cells (2×105 cells/well) were seeded into 6-well-plates and incubated until they reached a confluent monolayer. Before wound-creation, the cells were grown in serum-free DMEM/F12 medium for 24h. Subsequently, a 10-μL pipette tip was applied under sterile conditions to create an artificial wound on each plate. The cell debris was washed away with phosphate-buffered saline (PBS) after wound-creation, and the culture grown at 37°C in fresh DMEM/F12 medium for another 24h in a humidified atmosphere (5% CO2, 95% air). The average extent of wound closure was quantified. Experiments were completed independently in triplicate.
Cell Invasion and Migration Assay
The cell invasion and migration assays were performed using Transwell chambers (Corning, NY, USA) with or without pre-coated Matrigel (Corning, NY, USA), respectively. Cells (5×104 cells/well) were suspended in DMEM medium with 1% FBS and spread on the top transwell chamber. The chambers were incubated in a 24-well plate with culture medium supplemented with 10% FBS for 24h. The migrated or invasive cells were fixed in 4% PFA and stained with 0.1% crystal violet after careful removal of cells on the upper chamber surface. The cells were counted in five random fields of each filter. Experiments were independently conducted in triplicate.
Tumor Xenograft Experiments
Twelve BALB/c nude mice (4–6 weeks old, male) provided by Shanghai Laboratory Animal Research Center (Shanghai, China) were housed in specific pathogen-free (SPF) conditions. SCC9 cells (5×106) stably infected with either sh-ZEB1-AS1 or empty vector were subcutaneously injected into the armpit regions of nude mice under sterile conditions. The formula (V=width2 × length × 0.5) was used to calculate the volume of each xenograft weekly. Mice were sacrificed 28 days post-implantation and the xenograft tissues carefully dissected and weighed. Each tumor xenograft sample was embedded in optimal cutting temperature compound (OCT) and sliced into 5 μm sections for immunohistochemical (IHC) analysis.
IHC Analysis
The indirect streptavidin-peroxidase method was used for IHC analyses. Briefly, the endogenous peroxidase activity of tissue sections was eliminated with 3% H2O2 and blocked with a normal goat serum seal solution for 30 min at RT to prevent nonspecific binding. The tissues were incubated in anti-Ki67 primary antibodies overnight at 4°C. The next day, streptavidin-HRP-conjugated IgG (Beyotime, Shanghai, China) was applied to the sections prior to incubations at RT for another 2h, and the IHC reactions of tissues were detected using a DAB kit (Beyotime, Shanghai, China). Finally, the slides were observed under a phase contrast light microscope (Leica DMI6000B, Wetzlar, Germany). Image-Pro plus 6.0 software (Media Cybernetics, USA) was used to calculate the average integral optical density of each tissue section from images in five random fields.
Dual-Luciferase Reporter Assay
The direct interaction of ZEB1-AS1 and miR-23a was confirmed using the DLR assay. We constructed wild- and mutated-type ZEB1-AS1 vectors (WT-ZEB1-AS1, MUT-ZEB1-AS1) by cloning the lncRNA ZEB1-AS1 fragment into a psiCHECK2 vector (Promega, WI, USA). Co-transfection of recombinant plasmids (2ng) with either miR-23a (30 pmol) mimics or negative control (NC) was performed in 239T cells cultured in 24-well plates (6×104 cells/well) using Lipofectamine 3000 per manufacturer instructions (Invitrogen, CA, USA). The luciferase activity was measured with a Dual-Luciferase Assay Kit (Promega, WI, USA) after 48h transfection. Three independent experiments were conducted.
RT-qPCR Analysis
TRIzol (Invitrogen, CA, USA) was used to isolate total RNA from cells and tissues based on manufacturer protocols. The quality and quantity of RNA was determined with an ultraviolet spectrophotometer (Thermo Fisher Scientific, CA, USA). Reverse transcription was carried out using the High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, CA, USA). The cDNA template was amplified using the SYBR_PremixExTaq II kit (Takara, Dalian, China) on an ABI StepOne Real-Time PCR System (Applied Biosystems, CA, USA). For miRNA expression, the cDNA reverse transcription and qPCR were carried out with the PrimeScript® miRNA cDNA Synthesis kit (TaKaRa, Dalian, China) and the TaqMan Universal Master Mix II (Thermo Fisher Scientific, CA, USA), respectively. The expression levels of mRNAs or miRNAs were normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6 expression, respectively. The ΔCt or 2−ΔΔCt method was used to analyze the data. Table 2 displayed the primers used in our present study.
Table 2.
The Sequences of the Primers
| Gene Name | Sequence (5ʹ- > 3ʹ) |
|---|---|
| LncRNA ZEB1-AS1 | GCTCACGTGAAGTCCATC |
| CGACATCCTTGACGTTGCAC | |
| miR-23a-3p | GCGATCACATTGCCAGGG |
| CAGTGCGTGTCGTGGAGT | |
| vimentin | TGCCAACCGGAACAACGAT |
| AATTCTCTTCCATTTCACGCATC | |
| N-cadherin | CAAGATGGGTCAATGGAAATAG |
| CTCAGGAATACGAGCCTTCAC | |
| E-cadherin | GAGAACGCATTGCCACATACAC |
| AAGAGCACCTTCCATGACAGAC | |
| U6 | CTCGCTTCGGCAGCACA |
| AACGCTTCACGAATTTGCGT | |
| GAPDH | GCACCGTCAAGGCTGAGAAC |
| TGGTGAAGACGCCAGTGGA |
Western Blot Analysis
RIPA Lysis Buffer (Thermo Fisher Scientific, CA, USA) was used to isolate total proteins from cells or tissues. A BCA Protein Assay Reagent Kit (Beyotime, Shanghai, China) was used to measure protein concentration. Identical quantities of proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, MA, USA). The incubation of membranes with the primary antibodies, including anti-vimentin (1:2000), anti-N-cadherin (1:2000), and anti-E-cadherin (1:5000) was carried out overnight at 4°C after blocking with a 5% skim milk solution. The next day, the washed membranes were incubated in horseradish peroxidase-conjugated anti-rabbit secondary antibody. The antibodies in our present study were obtained from Abcam (Cambridge, USA). The ECL Western Blotting Kit (BioVision, CA, USA) was used to measure the protein bands on images taken by a ChemiDoc XRS system (Bio-rad, CA, USA).
Statistical Analysis
All data were presented as mean ± SD from at least three independent experiments. Comparisons were conducted using Student’s t-test (two-tailed) or one-way ANOVA as indicated. The differences among multiple groups were compared with the Kruskal–Wallis test (post-hoc Mann–Whitney U-test with Bonferroni’s). Kaplan-Meier survival curves were drawn with the Log rank test. P-values less than 0.05 were statistically significant in this study. The statistical analyses and plotting were performed with GraphPad Prism 6.0 software (GraphPad Software, CA, USA) and R software.
Results
Overexpression of ZEB1-AS1 in OSCC Tissues Was Linked to Poor Patient Prognosis
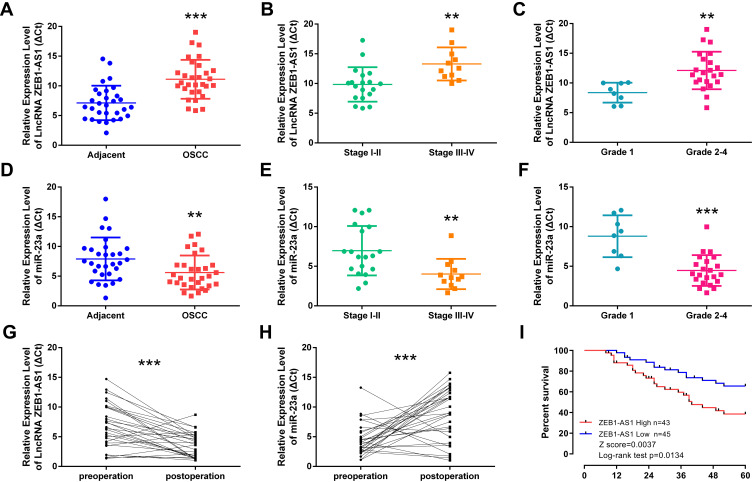
The lncRNA ZEB1-AS1 and miR-23a expression levels in 30 paired specimens (OSCC tissues and corresponding non-cancer tissues) were initially tested by qRT-PCR. The results showed that ZEB1-AS1 expression was increased and miR-23a expression levels were decreased in OSCC tissues compared to those in the corresponding non-cancer tissues (Figure 1A and D). To further explore ZEB1-AS1 effects on OSCC metastasis, the association between ZEB1-AS1 expression and clinicopathological features was analyzed. These results demonstrated that ZEB1-AS1 expression levels were remarkably higher, and that miR-23a levels were markedly lower, in OSCC tissues at advanced TNM stages (Stage III–IV) compared to an early stage (Stage I) (Figure 1B and E). The expression trend for ZEB1-AS1was similar in the pathological grade analysis (Figure 1C and F). Serum expression levels of ZEB1-AS1 were also detected by qRT-PCR, and the results showed that ZEB1-AS1 and miR-23a expression levels were decreased and increased, respectively, in serum after the resection of OC tissues (Figure 1G and H). Furthermore, the OS rates were lower in patients with high ZEB1-AS1 expression compared to those with low expression levels as shown by a Kaplan-Meier analysis (Figure 1I). These results suggest that the overexpression of ZEB1-AS1 may be involved in the pathogenesis of OSCC and linked to the poor prognosis of OSCC patients.
Figure 1.
ZEB1-AS1 overexpression in OSCC correlates with tumor progression and poor patient prognosis. (A, D) The relative expression of ZEB1-AS1 or miR-23a in OSCC and adjacent non-cancer tissues (n=30). (B, E) The relative expression of ZEB1-AS1 or miR-23a in OSCC tissues at different advanced stages of TNM. (C, F) The relative expression of ZEB1-AS1 or miR-23a in OSCC tissues at different pathology grades. (G, H) The relative expression of ZEB1-AS1 or miR-23a in the sera of OSCC patients pre- and post-operative. (I) Kaplan–Meier survival curve analysis of patients based on ZEB1-AS1 expression and OS rates. **P < 0.001; ***P < 0.0001.
Abbreviations: OSCC, Oral squamous cell carcinoma; OS, overall survival.
ZEB1-AS1 Promoted OSCC Cell Proliferation
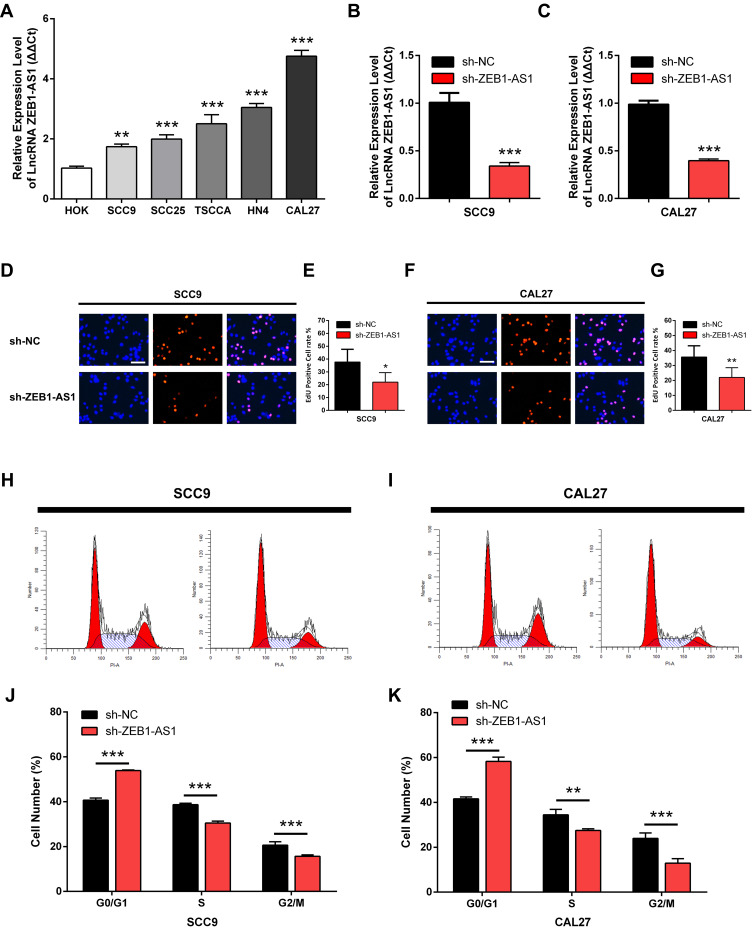
The expression levels of ZEB1-AS1 in OSCC cell lines were first determined by qRT-PCR to identify its roles in OSCC progression. The human normal oral epithelial cell line HOK was used as a control. The results showed that ZEB1-AS1 expression levels were markedly higher in all OSCC cells than in the HOK cell line (Figure 2A). SCC9 and CAL27 cell lines were used to explore the effect of ZEB1-AS1 on OSCC cell proliferation. We established ZEB1-AS1-silenced cell models by transfecting OSCC cells with the sh-ZEB1-AS1 plasmid. The expression of ZEB1-AS1 decreased in both SCC9 and CAL27 cells after sh-ZEB1-AS1 transfection, which was confirmed by qRT-PCRs (Figure 2B and C). Next, an EdU cell proliferation assay showed that the proliferative abilities of OSCC cells were suppressed by ZEB1-AS1 knockdowns (Figure 2D-G). We also found a reduced percentage of cells in the S phase and an increased percentage of cells in the G2M phase in SCC9 and CAL27 cells with a ZEB1-AS1 knockdown (Figure 2H-K). These findings suggest that lncRNA ZEB1-AS1 is a proliferative promoter in OSCC cells.
Figure 2.
ZEB1-AS1 knockdown inhibits OSCC cell proliferation in vitro. (A) The relative expression of ZEB1-AS1 in OSCC cell lines. (B, C) The expression of ZEB1-AS1 in OSCC cell lines following sh-ZEB1-AS1 transfection. (D–G) EdU assay in OSCC cell lines following ZEB1-AS1 knockdown. (H–K) The percentage of cells at S phase following ZEB1-AS1 knockdown. Data are presented as the means ± SD. *P < 0.05; **P < 0.001; ***P < 0.0001. Scale bar=50μm.
Abbreviation: OSCC, oral squamous cell carcinoma.
ZEB1-AS1 Promoted Migration and EMT in OSCC Cell Lines
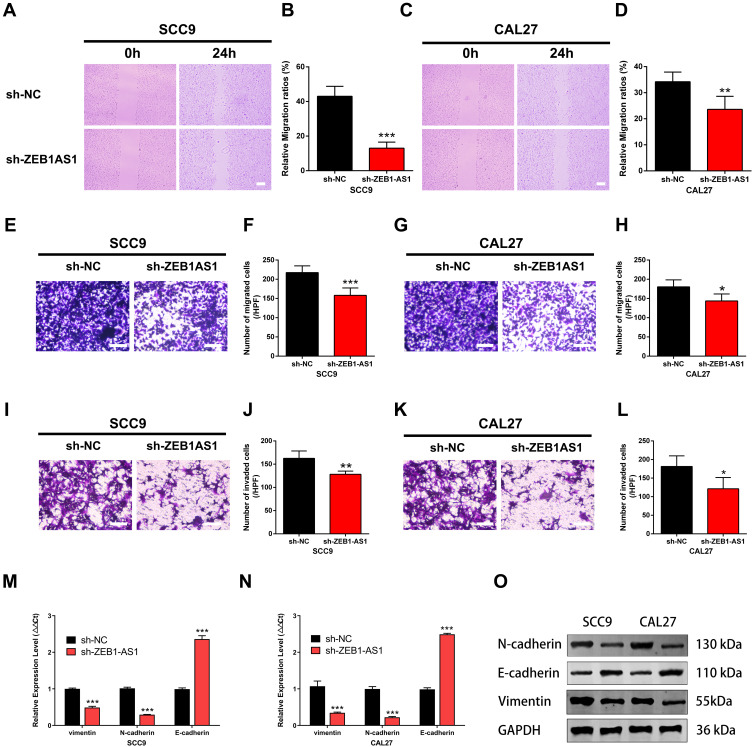
The wound healing, cell invasion, and cell migration assays were performed in SCC9 and CAL27 cells to further study the effects of ZEB1-AS1 on OSCC metastasis. The results showed that the healed area (Figure 3A-D) and the number of migrated cells (Figure 3E-H) were remarkably reduced in ZEB1-AS1-knocked down cells compared to negative controls. Next, we conducted an invasion assay to determine the role(s) of ZEB1-AS1 in the invasive capacities of OSCC cells. The invasion ability of SCC9 and CAL27 cells was dramatically suppressed by a ZEB1-AS1 knockdown compared to negative controls (Figure 3I-L). We further used qRT-PCR to test the expression levels of EMT-related genes (vimentin, N-cadherin, and E-cadherin). There was a marked reduction in the expression levels of vimentin and N-cadherin in addition to upregulated expression of E-cadherin after silencing ZEB1-AS1 (Figure 3M and N). Similar results were observed for the protein levels measured by Western blot analysis (Figure 3O). These findings demonstrate that OSCC metastasis can be facilitated by ZEB1-AS1-mediated promotion of cell invasion, migration, and EMT.
Figure 3.
ZEB1-AS1 knockdown inhibited OSCC cell migration and invasion. (A–D) The cell migration distance following sh-ZEB1-AS1 transfection. (E–H) The number of migrated cells following sh-ZEB1-AS1 transfection. (I–L) The number of invaded cells following sh-ZEB1-AS1 transfection. (M, N) The expression of EMT-related genes vimentin, N-cadherin, and E-cadherin following sh-ZEB1-AS1 transfection in OSCC cell lines. (O) The protein levels of vimentin, N-cadherin, and E-cadherin following sh-ZEB1-AS1 transfection in OSCC cell lines. Data are presented as the means ± SD. *P < 0.05; **P < 0.001; ***P < 0.0001. Scale bar=50μm.
Abbreviations: OSCC, oral squamous cell carcinoma; EMT, endothelial–mesenchymal transition.
ZEB1-AS1 Acted as a miR-23a Sponge in OSCC Cell Lines
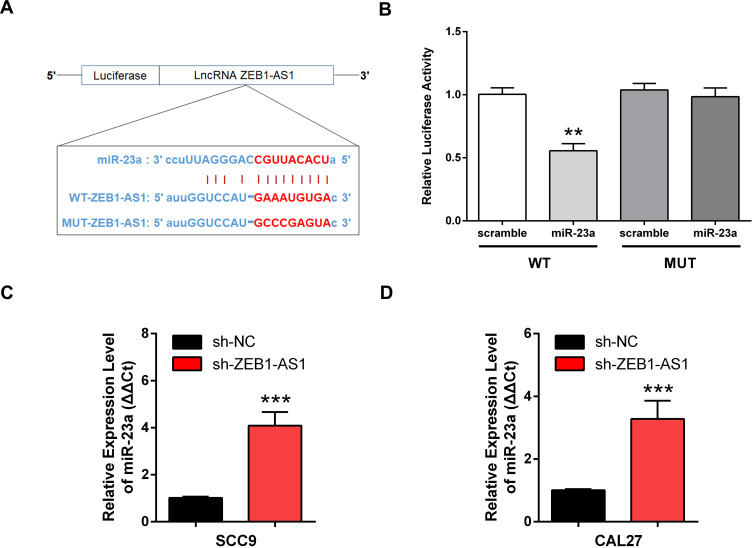
A putative miR-23a binding site was predicted by bioinformatics to be in the 3ʹ-UTR of ZEB1-AS1 and was used to study the mechanism(s) of ZEB1-AS1-mediated OSCC cell growth and metastasis (Figure 4A). Next, we conducted a DLR assay to evaluate the potential direct interaction between ZEB1-AS1 and miR-23a. The transfection of miR-23a mimics into WT-ZEB1-AS1 groups reduced the relative luciferase activity, whereas there was no change in the relative luciferase activity in the MUT-ZEB1-AS1 groups when transfected with miR-23a mimics (Figure 4B). Furthermore, miR-23a expression was increased by ZEB1-AS1 downregulation in OSCC cells (Figure 4C and D). These results indicated that ZEB1-AS1 interacted with and negatively regulated miR-23a in OSCC cells.
Figure 4.
ZEB1-AS1 acted as a miR-23a sponge in OSCC cell lines. (A) The predicted binding sites between miR-23a and ZEB1-AS1. (B) Luciferase activity in 293T cells measured using a DLR assay. (C, D) Expression of miR-23a in OSCC cell lines following ZEB1-AS1 knockdown. Data are presented as the means ± SD. **, P < 0.001; ***, P < 0.0001.
Abbreviations: OSCC, oral squamous cell carcinoma; DLR, dual-luciferase reporter.
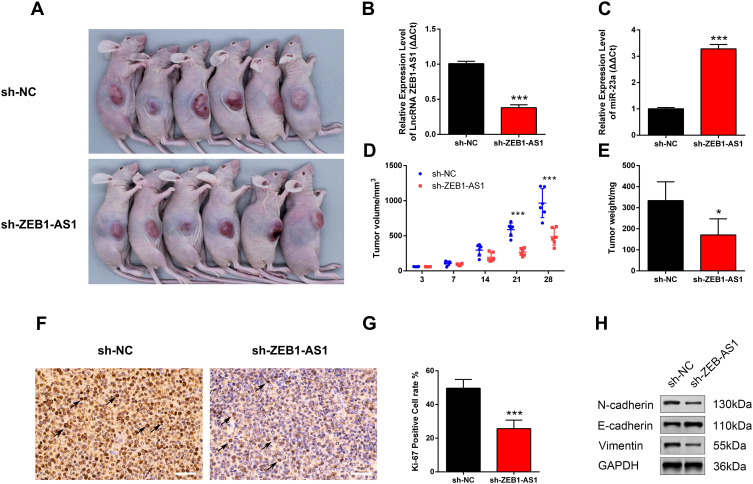
ZEB1-AS1 Suppressed Tumor Growth and Metastasis in Nude Mice
The nude mice xenograft tumor model was developed with SCC9 cells to study the roles of ZEB1-AS1 in tumorigenesis in vivo. All mice developed tumors after injection with SCC9 cells. The expression levels of ZEB1-AS1 and miR-23a were measured by qRT-PCR (Figure 5B and C). The volume of xenograft tumors was measured on days 3, 7, 14, 21, and 28 post-SCC9 cell injection. ZEB1-AS1 knockdown significantly reduced the tumor volumes when compared to tumors in the NC groups (Figure 5A and D). A similar result was obtained for tumor weights (Figure 5E). We also performed an IHC stain to detect tumor cell proliferation in vivo. The Ki-67-positive cell rate was reduced in the sh-ZEB1-AS1 groups compared to the NC groups (Figure 5F and G). Moreover, we confirmed reduced protein levels of N-cadherin and vimentin as well as increased E-cadherin expression in the ZEB1-AS1 knockdown groups (Figure 5H). These findings indicate that ZEB1-AS1 promoted OSCC metastasis by regulating cell proliferation and EMT progression.
Figure 5.
ZEB1-AS1 knockdown inhibited OSCC progression. (A) A gross look of xenograft tumors. (B, C) The relative expression of ZEB1-AS1 and miR-23a in OSCC xenograft tissues. (D) Tumor volumes were examined at days 3, 7, 14, 21, and 28 (n=6). (E) Tumor weights measured at day 28 (n=6). (F, G) Tumor cell proliferation was detected with IHC using an antibody against Ki-67. (H) The protein levels of vimentin, N-cadherin, and E-cadherin in OSCC xenograft tissues. Data are presented as the means ± SD. *, P < 0.05; ***, P < 0.0001. Scale bar=50μm.
Abbreviation: OSCC, oral squamous cell carcinoma.
Discussion
In our current study, the potential mechanisms of ZEB1-AS1 in OSCC growth and progression were elucidated. We first found that increased ZEB1-AS1 expression levels were present in both OSCC tumor tissues and cell lines, and that upregulated ZEB1-AS1 levels were associated with poor prognosis in OSCC patients. Moreover, the results of loss-of-function experiments demonstrated that the proliferation and EMT of OSCC cells could be suppressed by a ZEB1-AS1 knockdown in vitro. We also showed that tumor growth and metastasis can be suppressed by the knockdown of ZEB1-AS1 in vivo. Mechanistically, we demonstrated that ZEB1-AS1 is a sponge for miR-23a in OSCC cells. These results demonstrate that ZEB1-AS1 could be used as a biomarker for oral cancer diagnosis, and is promising for the development of biotherapeutics for the clinical treatment of this dangerous malignancy.
In human genome, more than 98% of RNA sequences are defined as non-coding RNAs (ncRNAs) due to their inability to encode proteins. LncRNAs are the main components of ncRNAs, accounting for about 68% of total lncRNAs.22,23 Increasing evidence indicates that lncRNAs may participate in diverse cellular processes such as cell cycle progression, proliferation, migration, invasion, and chemoresistance.24,25 Therefore, the regulatory mechanisms of lncRNAs should be thoroughly explored for their potential to develop promising novel strategies to treat multiple diseases. The molecular features and functions of lncRNAs in cancer progression have been investigated.26,27 However, the expression levels and specific roles of lncRNAs in cancers including OSCC remain unclear. The expression levels and functions of lncRNAs may be vastly different among cell types due to their strong tissue specificity.28 This characteristic could lead to completely different biological functions of lncRNA on tumor growth and metastasis in different types of cancers. For instance, Huang et al reported that lncRNA- NEAT1, acting as a ceRNA of miR-365, could induce cell cycle arrest in G0/G1 phase as well as promote cell proliferation and invasion by regulating the target RGS20 in OSCC cells.29 Wang et al demonstrated that delivery of lncRNA-H19 increased AMPKα expression and subsequently reduced MMP9 activation, which suppressed tumor metastasis in gastric cancer.30
Considerable evidence indicates that lncRNA ZEB1-AS1 participates in diverse types of human cancers. Ni et al found that colon adenocarcinoma cell growth and metastasis could be facilitated by ZEB1-AS1 through competitive binding with miR-455-3p, which results in PAK2 overexpression induced by the PAK2 mRNA transcripts releasing miR-455-3p.31 Qu et al reported that non-small cell lung cancer (NSCLC) progression was linked to the ZEB1-AS1/miR-409e-3p/ZEB1 feedback loop.32 However, the exact mechanism for ZEB1-AS1 promotion of OSCC metastasis is still unknown. To investigate the role of ZEB1-AS1 in OSCC progression, we measured the expression of ZEB1-AS1 in five different OSCC cell lines. These oral cancer cell lines originated from tongue squamous cell carcinoma tissue or laryngeal squamous cell carcinoma tissue, respectively. Previous studies demonstrated that different oral cancer cell lines have their own biological characteristics and gene expression profiles. Confirm the aberrant ZEB1-AS1 expression in various oral cancer cell lines makes our results more convincing, and our results showed that all oral cancer cell lines used in the current study have a higher ZEB1-AS1 expression when compared with normal oral epithelial cell line HOK. Next, the cell lines SCC9 (lncRNA ZEB1-AS1 low expression) and CAL27 (lncRNA ZEB1-AS1 high expression) were chosen for functional studies. The malignant phenotypes, tumor cell invasion and migration EMT could promote distant metastases of tumors, and accumulating studies indicate that lncRNAs play important roles in this process. In our present study, the protein expression levels of EMT-related genes were measured by Western blot analysis to determine the potential for ZEB1-AS1-mediated EMT in OSCC cells. The results showed that considerable N-cadherin, vimentin downregulation as well as E-cadherin overexpression were caused by ZEB1-AS1 knockdowns both in vitro and in vivo. These findings suggest that OSCC cells may be promoted by ZEB1-AS1 activity that promotes EMT.
Regarding the molecular mechanism, lncRNAs commonly exert their functions in tumorigenesis and progression through interactions with miRNAs and modulating expression of miRNA downstream targets. To identify the ZEB1-AS1 ceRNA network in OSCC cells, we used a bioinformatics analysis to identify a candidate miRNA, miR-23a. Our data showed that miR-23a expression levels were related to ZEB1-AS1 expression in OSCC clinical tissues and cell lines. Furthermore, we found that ZEB1-AS1 directly binds to miR-23a in OSCC cells, which was detected by a luciferase reporter assay. A limitation to this study was the lack of functional studies on ZEB1-AS1 and miR-23a, and further experimental models are warranted to confirm these results.
In summary, ZEB1-AS1 upregulation was correlated with aggressive OSCC phenotypes and a poor prognosis in patients with OSCC. The proliferation, cell cycle progression, invasion, migration, and EMT of OSCC cells can be promoted by ZEB1-AS1 expression. These results provide new insights into the regulatory effects of ZEB1-AS1 on the progression of oral cancer and demonstrate that ZEB1-AS1 may potentially be used for the diagnosis and treatment of OSCC.
Acknowledgments
We would like to thank TopEdit (www.topeditsci.com) for English language editing of this manuscript.
Funding Statement
The work was funded by National Natural Science Foundation of China (81972535).
Abbreviations
OSCC, oral squamous cell carcinoma; DLR, dual-luciferase reporter; EMT, epithelial–mesenchymal transition; TNM, tumor node metastasis.
Data Sharing Statement
The datasets and supporting materials generated and/or analyzed during the current study are available from the corresponding author upon a reasonable request.
Ethics Approval and Consent to Participate
Written informed consent was obtained from all patients. The study protocol was approved by the Ethics Committee of China Medical University and performed in accordance with the tenets of the Declaration of Helsinki. The mice were bred in the animal experimental center following procedures approved by the Institutional Animal Care and Use Committee of China Medical University and cared for in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Habbous S, Harland LT, La Delfa A, et al. Comorbidity and prognosis in head and neck cancers: differences by subsite, stage, and human papillomavirus status. Head Neck. 2014;36(6):802–810. doi: 10.1002/hed.23360 [DOI] [PubMed] [Google Scholar]
- 2.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982 [DOI] [PubMed] [Google Scholar]
- 3.Adoch W, Garimoi CO, Scott SE, et al. Knowledge of cervical cancer risk factors and symptoms among women in a refugee settlement: a cross-sectional study in northern Uganda. Confl Health. 2020;14(1):85. doi: 10.1186/s13031-020-00328-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zini A, Czerninski R, Sgan-Cohen HD. Oral cancer over four decades: epidemiology, trends, histology, and survival by anatomical sites. J Oral Pathol Med. 2010;39(4):299–305. doi: 10.1111/j.1600-0714.2009.00845.x [DOI] [PubMed] [Google Scholar]
- 5.Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma–an update. CA Cancer J Clin. 2015;65(5):401–421. doi: 10.3322/caac.21293 [DOI] [PubMed] [Google Scholar]
- 6.Takahashi H, Yanamoto S, Yamada S, et al. Effects of postoperative chemotherapy and radiotherapy on patients with squamous cell carcinoma of the oral cavity and multiple regional lymph node metastases. Int J Oral Maxillofac Surg. 2014;43(6):680–685. doi: 10.1016/j.ijom.2013.11.013 [DOI] [PubMed] [Google Scholar]
- 7.Khanduja JS, Calvo IA, Joh RI, Hill IT, Motamedi M. Nuclear noncoding RNAs and genome stability. Mol Cell. 2016;63(1):7–20. doi: 10.1016/j.molcel.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 9.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li GY, Wang W, Sun JY, et al. Long non-coding RNAs AC026904.1 and UCA1: a “one-two punch” for TGF-beta-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics. 2018;8(10):2846–2861. doi: 10.7150/thno.23463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni W, Zhang Y, Zhan Z, et al. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol. 2017;10(1):91. doi: 10.1186/s13045-017-0449-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao S, Zhao ZY, Wu R, Zhang Y, Zhang ZY. Prognostic value of long noncoding RNAs in gastric cancer: a meta-analysis. Onco Targets Ther. 2018;11:4877–4891. doi: 10.2147/OTT.S169823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Wang N, Zhang X, Liu H, Yang S. LncRNA ZEB1-AS1 down-regulation suppresses the proliferation and invasion by inhibiting ZEB1 expression in oesophageal squamous cell carcinoma. J Cell Mol Med. 2019;23(12):8206–8218. doi: 10.1111/jcmm.14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma ZJ, Wang Y, Li HF, et al. LncZEB1-AS1 regulates hepatocellular carcinoma bone metastasis via regulation of the miR-302b-EGFR-PI3K-AKT axis. J Cancer. 2020;11(17):5118–5128. doi: 10.7150/jca.45995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng R, Li N, Yang S, Liu L, Han S. Long non-coding RNA ZEB1-AS1 promotes cell invasion and epithelial to mesenchymal transition through inducing ZEB1 expression in cervical cancer. Onco Targets Ther. 2018;11:7245–7253. doi: 10.2147/OTT.S179937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Kim SJ, Choo J. miR-23a-3p is a key regulator of IL-17C-induced tumor angiogenesis in colorectal cancer. Cells. 2020;9(6):1363. doi: 10.3390/cells9061363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin L, Xu G, Zhu Y, Wang Y. Expression of miR-23a and miR-135 and tumor markers in gastric cancer patients and the significance in diagnosis. Oncol Lett. 2019;18(6):5853–5858. doi: 10.3892/ol.2019.10943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu P, Wang C, Ma C, Wu Q, Zhang W, Lao G. MicroRNA-23a regulates epithelial-to-mesenchymal transition in endometrial endometrioid adenocarcinoma by targeting SMAD3. Cancer Cell Int. 2016;16(1):67. doi: 10.1186/s12935-016-0342-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koller K, Das S, Leuschner I, Korbelius M, Hoefler G, Guertl B. Identification of the transcription factor HOXB4 as a novel target of miR-23a. Genes Chromosomes Cancer. 2013;52(8):709–715. doi: 10.1002/gcc.22066 [DOI] [PubMed] [Google Scholar]
- 20.He Y, Meng C, Shao Z, Wang H, Yang S. MiR-23a functions as a tumor suppressor in osteosarcoma. Cell Physiol Biochem. 2014;34(5):1485–1496. doi: 10.1159/000366353 [DOI] [PubMed] [Google Scholar]
- 21.Siegel C, Li J, Liu F, Benashski SE, McCullough LD. miR-23a regulation of X-linked inhibitor of apoptosis (XIAP) contributes to sex differences in the response to cerebral ischemia. Proc Natl Acad Sci USA. 2011;108(28):11662–11667. doi: 10.1073/pnas.1102635108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9(3):e1471. doi: 10.1002/wrna.1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18(1):206. doi: 10.1186/s13059-017-1348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, Pan LM, Chen LB, Wang Y. LncRNA XIST promotes human lung adenocarcinoma cells to cisplatin resistance via let-7i/BAG-1 axis. Cell Cycle. 2017;16(21):2100–2107. doi: 10.1080/15384101.2017.1361071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu P, Chen X, Xie R, et al. lncRNA HOXD-AS1 regulates proliferation and chemo-resistance of castration-resistant prostate cancer via recruiting WDR5. Mol Ther. 2017;25(8):1959–1973. doi: 10.1016/j.ymthe.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding L, Ren J, Zhang D, et al. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis. 2018;39(3):397–406. doi: 10.1093/carcin/bgy006 [DOI] [PubMed] [Google Scholar]
- 27.Fang Z, Zhao J, Xie W, Sun Q, Wang H, Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by suppressing miR-184 expression. Cancer Med. 2017;6(12):2897–2908. doi: 10.1002/cam4.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Jiang Z, Wang S, et al. Long non-coding small nucleolar RNA host genes in digestive cancers. Cancer Med. 2019;8(18):7693–7704. doi: 10.1002/cam4.2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang G, He X, Wei XL. lncRNA NEAT1 promotes cell proliferation and invasion by regulating miR365/RGS20 in oral squamous cell carcinoma. Oncol Rep. 2018;39(4):1948–1956. doi: 10.3892/or.2018.6283 [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Yang X, Kai J, et al. HIF-1alpha-upregulated lncRNA-H19 regulates lipid droplet metabolism through the AMPKalpha pathway in hepatic stellate cells. Life Sci. 2020;255:117818. doi: 10.1016/j.lfs.2020.117818 [DOI] [PubMed] [Google Scholar]
- 31.Ni X, Ding Y, Yuan H, et al. Long non-coding RNA ZEB1-AS1 promotes colon adenocarcinoma malignant progression via miR-455-3p/PAK2 axis. Cell Prolif. 2020;53(1):e12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu R, Chen X, Zhang C. LncRNA ZEB1-AS1/miR-409-3p/ZEB1 feedback loop is involved in the progression of non-small cell lung cancer. Biochem Biophys Res Commun. 2018;507(1–4):450–456. doi: 10.1016/j.bbrc.2018.11.059 [DOI] [PubMed] [Google Scholar]