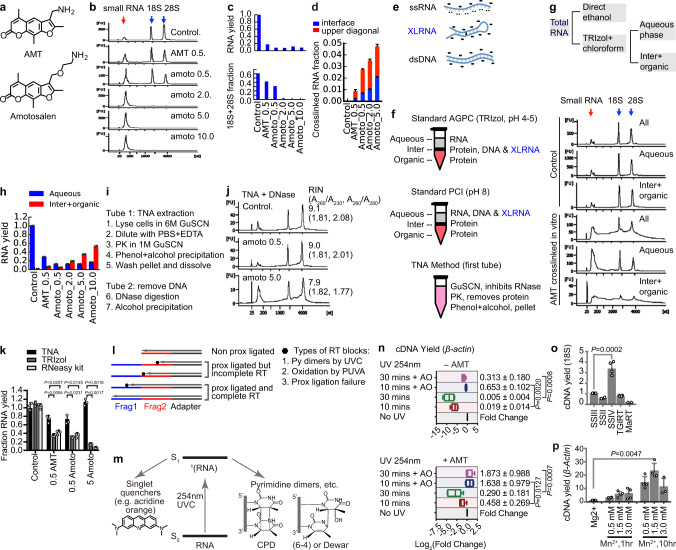
Fig. 2. Systematic optimization of PARIS2.
a Structures of AMT and amotosalen. b Higher concentrations of crosslinkers increase crosslinking efficiency. Psoralen crosslinked HEK293 cells RNA was extracted using the standard AGPC (TRIzol) method. Small RNA: RNA in the range of 50–300 nt, including tRNA, snRNA, 5S and 5.8S rRNAs etc. c Total RNA yield (upper panel) and 18S + 28S percentage (lower panel) from panel (b). d Fraction of psoralen crosslinked RNA from DD2D gels. Data are mean ± s.d.; n = 2 biological replicates. e Charge and hydrogen bonding of RNA and DNA molecules in standard AGPC (TRIzol) purification. The ‘-’ indicates negative charges. The ‘..’ indicates exposed bases involved in hydrogen bonding. f Phase partition of DNA, RNA and protein in AGPC and PCI methods, and comparison to TNA. g Repartition of crosslinked RNA to the interphase in Trizol extraction. Purified total RNA was treated with or without 365 nm UV plus AMT, and then either directly precipitated using ethanol, or extracted from the 2 phases of TRIzol-chloroform. h Recovery of crosslinked RNA from the aqueous and inter+organic phases using the TNA method. Data are mean ± s.d.; n = 2 biological replicates. i TNA method outline. j Recovery of inact RNA from psoralen crosslinked HEK293 cells using TNA method, after DNase treatment. RNA integrity numbers (RIN) and two indicators of RNA quality, A260/A230 and A260/A280, were listed. k Quantity of RNA isolated from control and AMT/amotosalen crosslinked cells. Data are mean ± s.d.; n = 3 biological replicates. two-tailed, unpaired t-test. l Effects of UV damages and failed proximity ligations (non prox ligated) on the yield of gapped reads. m UVC excited RNA form various products, such as pyrimidine dimers, hydrates and strand breaks. Alternatively, the energy can be transferred to singlet quenchers like acridine orange, leaving RNA intact. S0 and S1: ground state and excited singlet state. n cDNA yield from RNA irradiated with 254 nm UV, with or without AO protection, normalized to non-photo-reversal sample. Top: Non-crosslinked. Bottom: AMT crosslinked. Box plots show center line as median, box limits as upper and lower quartiles, whiskers as minimum to maximum values. 10 min + AO: n = 4; others: n = 7, biological replicates. two-tailed, unpaired t-test. o cDNA yield obtained in RT-qPCR experiments for PUVA damaged 18S-rRNA, normalized to SSIII. MaRT: Marathon RT. Data are mean ± s.d.; SSIII/SSIV/TGIRT: n = 4; SSII/MaRT: n = 2, biological replicates. two-tailed, unpaired t-test. p cDNA yield for β-Actin using SSIV in different reaction buffers and different incubation time, normalized to a standard Mg2+ condition. Data are mean ± s.d.; n = 4 biological replicates. two-tailed, unpaired t-test.