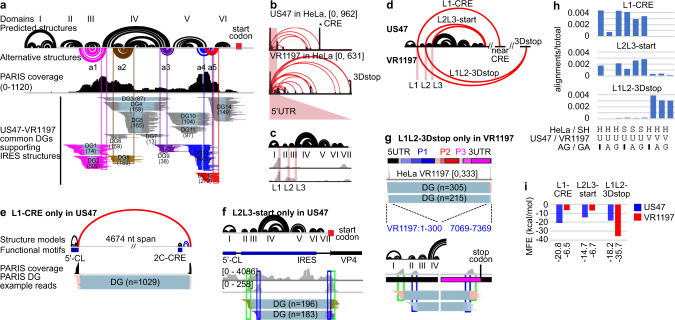
Fig. 6. PARIS2 reveals dynamic and long-range structures in the 5′UTR of two EV-D68 strains.
a PARIS2 validates predicted structures (black arcs) and reveals new alternative conformations (colored arcs) in the 699 nt 5′UTR of the US47 strain of EV-D68. 5′CL cloverleaf structure, IRES internal ribosome entry site, including domains II–VI, a1–a5 five alternative conformations. Numbers in parentheses are the counts of gapped alignments in each DG. b Long-range structures connecting the 5′UTR to the rest of the genome in two EV-D68 strains. All DGs have cov ≥ 0.01 for these two samples. The two stars indicate the right-side anchors for the strongest long-range strucgtures in the two strains, near the CRE and the region near the stop codon at the end of peptide 3D (3Dstop). c Long-range structures in the 5′UTR are primarily anchored in three linker regions L1–L3 (e.g., L1 is after domain I). d Diagram showing the three strongest long-range structures L1-CRE (connecting L1 and near CRE, in US47), L2L3-start (start codon, in US47), and L1L2-3Dstop (in VR1197). e–g DGs supporting the three long-range structures. In L1-CRE (e), the first ~5000 nt of the genome is shown. In the L2L3-start (f), the first ~800 nt of the genome is shown. In L1L2-3Dstop (g), the entire genome is shown. h Quantifying the strength of DGs for the three structures in all conditions. H HeLa cells, S SH-SY5Y cells, U US47, V VR1197, A AG, G GA. i Stability of the three structures, measured in minimal free energy (MFE), in the two strains.