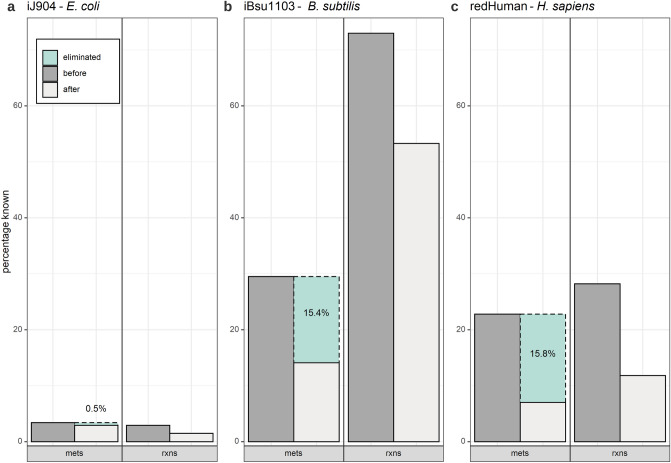
Figure 2.
Application of lumping on genome-scale metabolic models. The percentage of reactions with unknown is in three GEMs, (a) iJR904, (b) iBsu1103, and (c) redHuman, reduced after lumping due to elimination of metabolites with unknown . Shown is the percentage of unknown -values in relation to the total number of present metabolites (mets) and reactions, respectively, (rxns) before and after the lumping procedure was applied. The number of eliminated metabolites with unknown is with respect to the total number of metabolites. The investigated genome-scale models have low, high and medium number of unknown and , respectively, and pertain to organisms of increasing complexity.