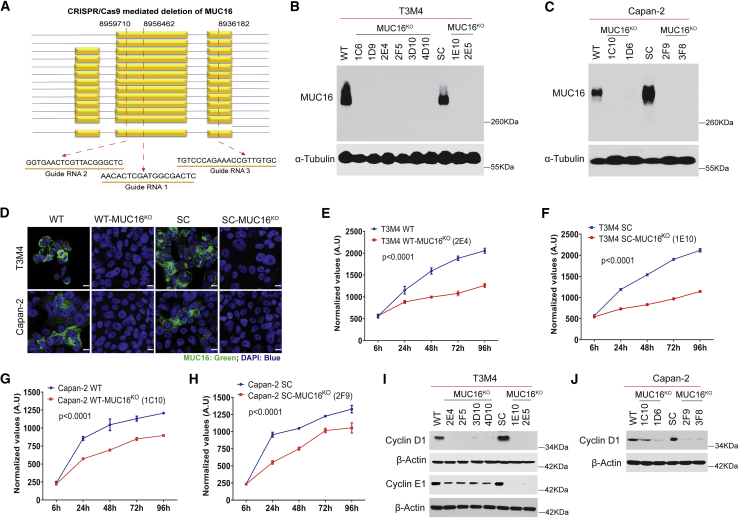
Figure 3.
Genetic deletion of MUC16 in PDAC cells
(A) Schematic representation of the targeted deletion of MUC16 via CRISPR-Cas9 constructs in T3M4 and Capan-2 (WT and SC) cells with three gRNA target loci are shown as red dotted lines. (B and C) Western blotting of MUC16 in T3M4 and Capan-2 (WT and SC) parental cells and MUC16KO clones respectively. Detection of α-tubulin served as a loading control. (D). Immunofluorescence analysis of MUC16 in T3M4 WT, WT-MUC16KO (2E4), T3M4 SC, SC-MUC16KO (1E10), Capan-2 WT, WT-MUC16KO (1C10), Capan-2 SC, and SC-MUC16KO (2F9) cells. Scale bar, 10 μm. (E–H) Cell proliferation assays in T3M4 WT, WT-MUC16KO (2E4) (E), T3M4 SC, SC-MUC16KO (1E10) (F), Capan-2 WT, WT-MUC16KO (1C10) (G), and Capan-2 SC, SC-MUC16KO (2F9) (H) cells. Data were presented as mean ± SD (n = 6; Tukey’s multiple comparisons test). (I) Western blotting of Cyclin D1 and Cyclin E1 in T3M4 WT, WT-MUC16KO clones, SC, and SC-MUC16KO clones. (J) Western blotting of Cyclin D1 in Capan-2 WT, WT-MUC16KO clones, SC, and SC-MUC16KO clones. β-actin was used as a loading control.