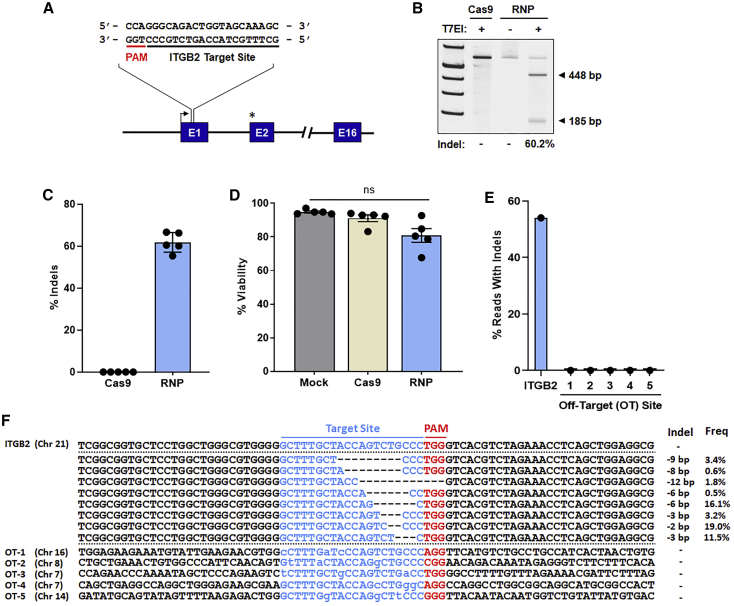
Figure 2.
Cas9 target site validation
(A) Schematic of the sgRNA/Cas9 RNP target site in non-coding E1 of the ITGB2 gene. The TSS is represented by a black arrow. The start codon within E2 is indicated by an asterisk. (B) Representative gel image of the T7 endonuclease I (T7EI) cleavage assay for quantification of sgRNA/Cas9 RNP-mediated indels 4 days post-electroporation (EP). Expected amplicon sizes are indicated. (C) Indel frequencies as measured by T7E1 cleavage assay (n = 5 independent donors). (D) Viability of mock-treated (electroporated only), Cas9-treated, and sgRNA/Cas9 RNP-treated cells 4 days post-EP (n = 5 independent donors). (E and F) Off-target analysis of the ITGB2 sgRNA/Cas9 RNP complex. Shown are percent sequencing reads with indels (E) and summary of individual indel size and frequency (F) at the ITGB2 on-target site and five in silico-predicted off-target (OT) sites. Total indel frequencies quantified by T7EI assay (C) and deep sequencing (E) were comparable at the ITGB2 on-target site. No indels were observed at the selected OT sites. Sequences for each target site are displayed in blue font; for OT sites, bases differing from the ITGB2 target sequence are indicated by lowercase letters. Respective protospacer adjacent motif (PAM) sequences (NGG) are highlighted in red font. Dashes represent deleted bases. In (C) and (D), results are displayed as mean ± SEM; ns, not significant by one-way repeated-measures ANOVA tests.