Figure 4.
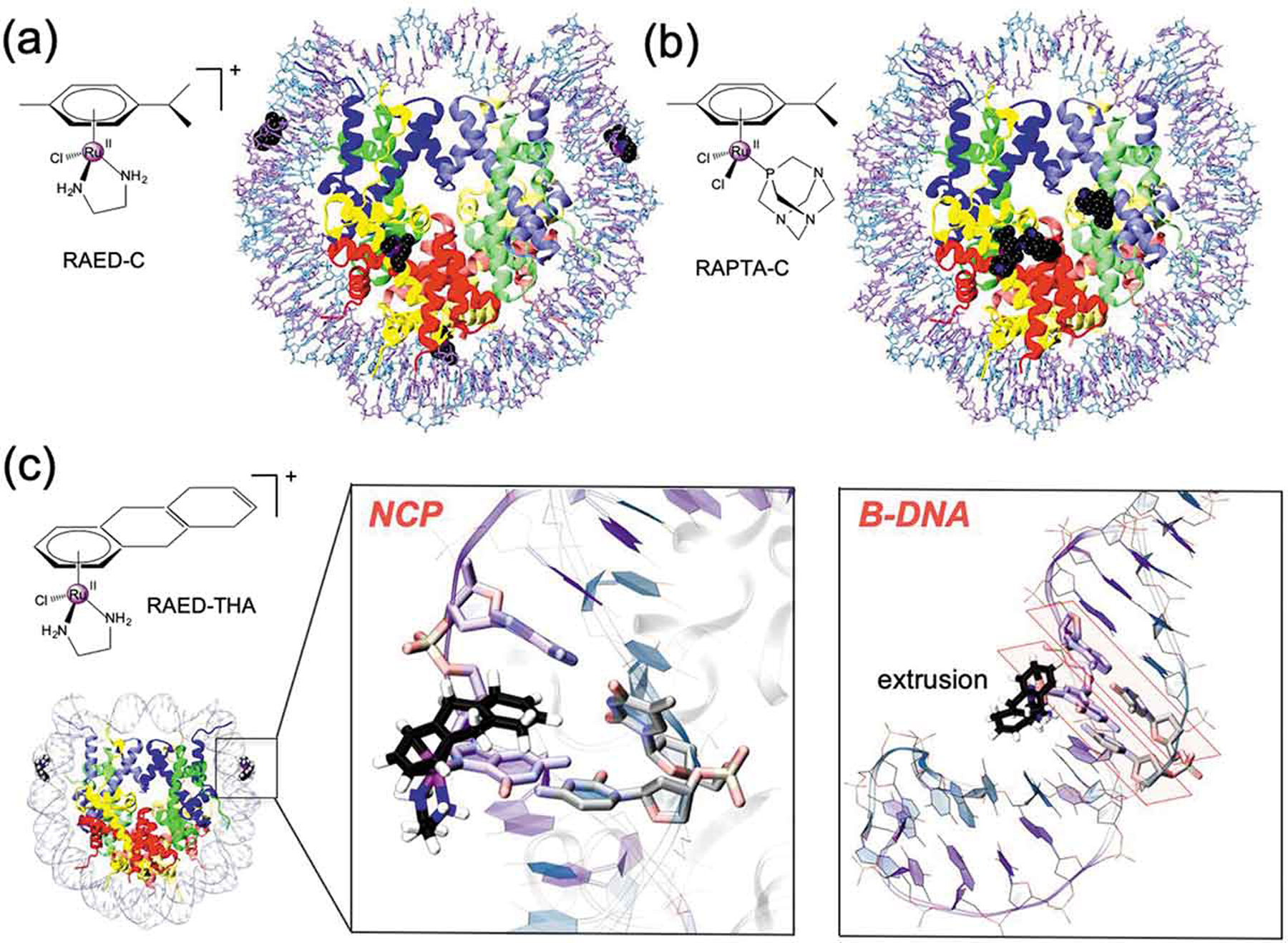
Chemical structure and binding mode of the RAED-C (A), RAPTA-C (B) and RAED-THA (C) at the Nucleosome Core Particle (NCP). RAED-C preferentially binds at DNA, while RAPTA-C selects protein histones. RAED-THA binds at the SHL ±1.5 sites of the DNA and engages ligand coordination with a guanine base (i.e. G ± 15). Within the nucleosomal DNA (close-up view), RAED-THA intercalates within the ±15/16 AG = CT step via a ‘mono-base stacking’ mechanism, which directly involves G ± 15. (B) Selected snapshots from classical MD simulations of a 14-mer DNA extracted from the RAED-THA/NCP model. After a ~ 20/30 ns MD, the THA is extruded from the double strand, which assumes a B-configuration. The typical base-stacking of DNA in B-configuration is highlighted in orange.
