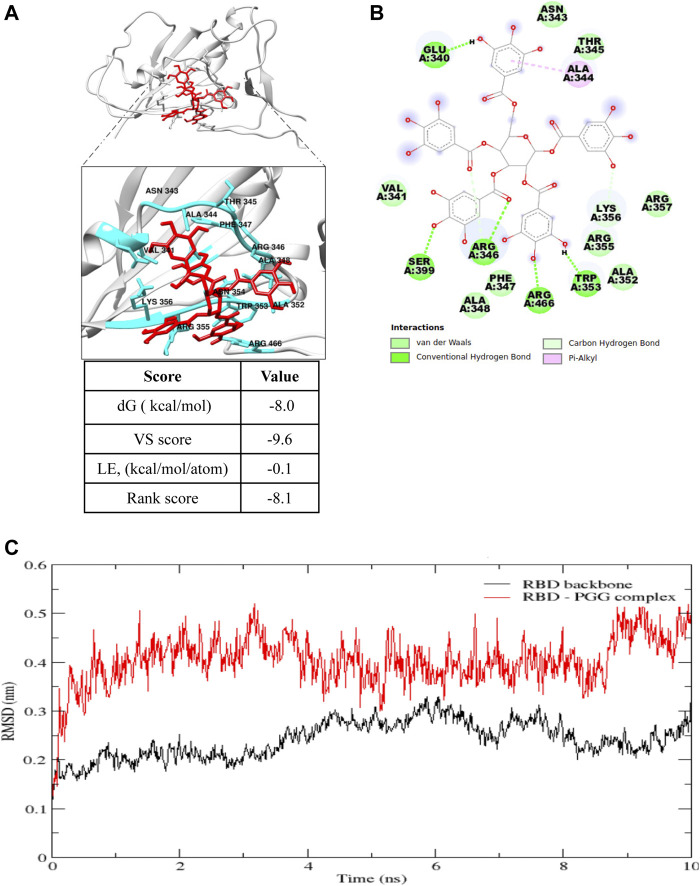
FIGURE 1.
Computational docking prediction of 1,2,3,4,6-Pentagalloyl glucose (PGG). (A) Molecular docking result showing the best binding pose of interaction, and (B) the residues involved and types of interaction of PGG with the receptor binding domain of the SARS-CoV-2 spike protein. (C) Root mean square deviation (RMSD) plot of the SARS-CoV-2 spike protein receptor binding domain (RBD) backbone alone and in complex with PGG during the 10 ns molecular dynamics simulation.