Extended Data Fig. 3. Binding and neutralizing activity of mAbs to SARS-CoV-2 variants.
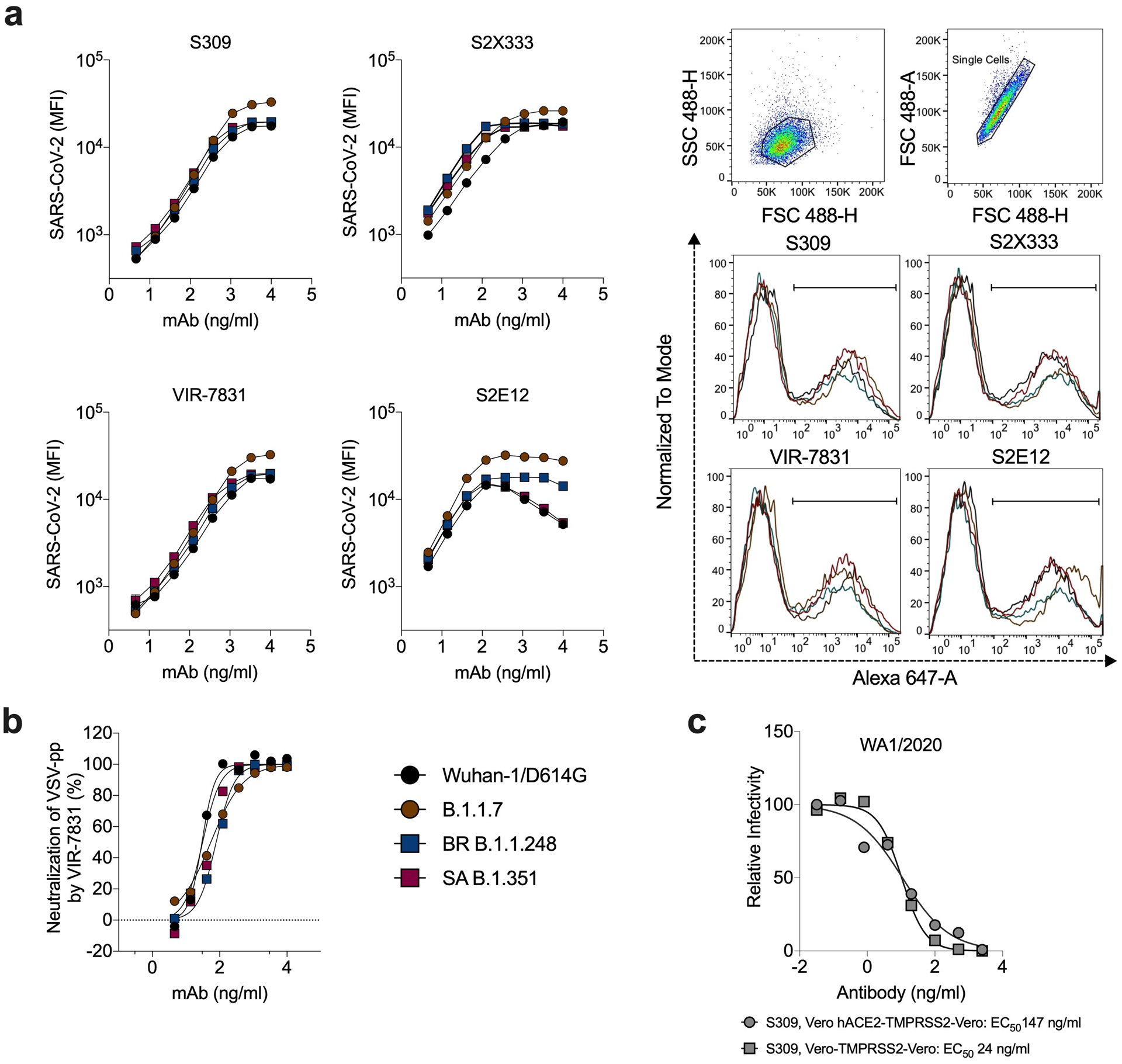
a, Binding of mAbs S2E12 (class 1, RBM), S309 (class 2, RBD base), VIR-7381 (class 2, RBD-base), and S2X333 (class 3, NTD) to SARS-CoV-2 spike proteins from the indicated strains when expressed on the surface of ExpiCHO cells (symbols, mean of duplicates from one experiment). Gating strategy and binding of mAbs S2E12, S309, VIR-7831, and S2X333 (at 370 ng/mL) to SARS-CoV-2 spike from indicated variants when expressed on the surface of ExpiCHO cells. Shown on histograms is the gating strategy of the population of positive cells (percentage ranging between 37 and 46%) used to calculate MFI. b, Neutralization of VSV-SARS-CoV-2 pseudotyped viruses (with indicated spike proteins) on Vero E6 cells. Mean ± standard deviation of sextuplicates is shown for all pseudoviruses, except for SARS-CoV-2 WT (mean of triplicates). WT, Wuhan-1 + D614G. c, Serial dilutions of S309 mAb were mixed with Vero CCL81 cell-derived WA1/2020 and added to Vero-hACE2-TMPRSS2 or Vero-TMPRSS2 cells for evaluation of neutralizing activity by FRNT. One representative experiment of two is shown. The EC50 values are provided in the legend in ng/mL.
