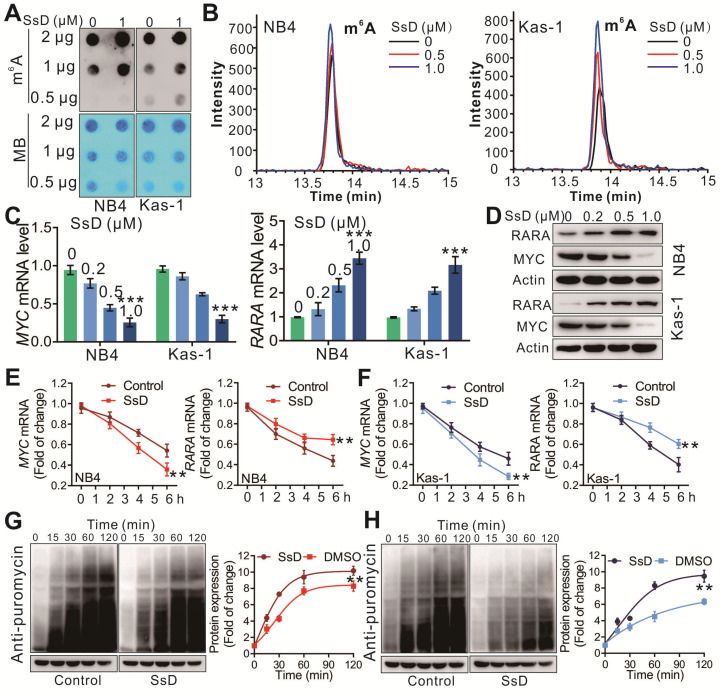
Figure 4.
SsD induces m6A modification. (A) m6A dot blots of indicated cells treated with SsD are shown. MB (methylene blue) represents the loading control of RNA samples. (B) TSQ traces of FTO demethylation of m6A in ssRNA in the absence and presence of the SsD in NB4 and Kas-1 cells are shown, respectively. (C) Expression levels of indicated genes in cells treated with SsD measured using qPCR. Data represent three independent experiments. (D) Western blot analysis of the protein levels of indicated genes in SsD treated cells. (E-F) qPCR analysis of NB4 and Kas-1 cells treated with SsD for 24 h, followed by 5 µg/mL actinomycin-D treatment for the indicated time points. Gene expression was normalized to GAPDH. (G-H) Western blot analysis for protein stability in cells treated with SsD for 24 h, followed by 200 nM puromycin treatment for the indicated time points. Graphs are showing the quantification of Western blot analysis normalized to β-actin; Data represent three independent experiments and present the mean ± SD; *p < 0.05, **p < 0.01, ***p < 0.001.