Figure 9.
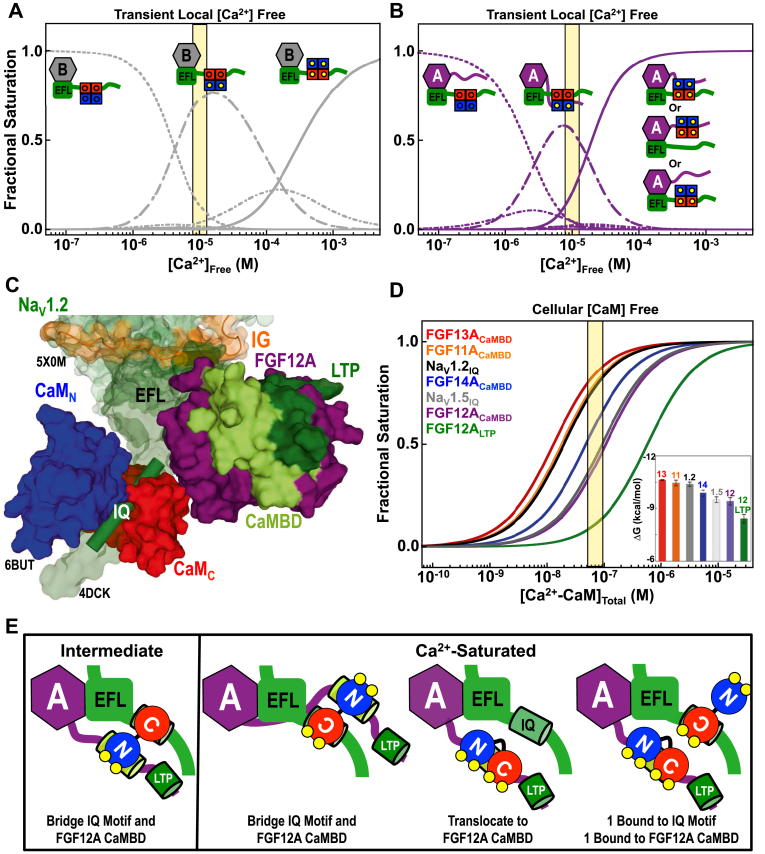
States of CaM bound to FGF12B+NaV1.2CTDor FGF12A+NaV1.2CTD.A and B, abundance of ligated CaM species in the CaM+FGF12B+NaV1.2CTD (A, gray) and CaM+FGF12A+NaV1.2CTD (B, purple) complexes. Simulations are based on the Ca2+-binding affinities reported in Table 4. States of CaM with an abundance >0.25 are shown in the inset schematics that depict the possible orientations of the CaM domains (CaMN/blue, CaMC/red) relative to FGF12B (gray) and NaV1.2CTD (green) (A) or FGF12A (purple) and NaV1.2CTD (green) (B), Ca2+ are shown as yellow circles. The yellow box is centered at physiologically relevant Ca2+ concentration of 10 μM. Representing the occupancy of the four Ca2+-binding sites in CaM (in order I, II, III, IV) with binary, where 0 is emtpy and 1 if filled, the discernible (abundances >0.05) species of CaM in the CaM+FGF12B+NaV1.2CTD complex (A) are 0000 (dotted line), 1100 (dashed and dotted line), 1101 or 1110 (dashed line), and 1111 (solid line). The discernible (abundance >0.5) species of CaM in the CaM+FGF12A+NaV1.2CTD complex (B) are 0000 (dotted line), 1000 or 0100 (dashed line), 1100 (dashed and dotted line), 1111 (solid line). C, model of CaM (CaMN/blue, CaMC/red) bound to an NaV CTD (forest green, surface) relative to the NaV DIII-DIV linker (orange) and FGF12A (purple, LTP/green, CaMBD/limon). The model is a composite of NaV1.5 CTD (4DCK), aligned with NaVPAS (5X0M) EFL (a.a. 1426–1521). NaVPAS a.a. 1122–1160 are shown as the DIII–DIV linker. The apo CaM+NaV1.2IQp ensemble (6BUT) was aligned with 4DCK via CaM a.a. 101–112 and 117–128. The NaV1.2IQp (a.a. 1904–1924) is shown as a forest green cylinder. The model of FGF12A was aligned with 4DCK using FGF13U a.a. 11–158. For clarity CaM and FGF13B in 4DCK are hidden. D, simulation of the saturation of each A-type FGF CaMBD (FGF11A/orange, FGF12A/purple, FGF13A/red, FGF14A/blue), FGF12A LTP (green) or the IQ motifs of NaV1.2 (black) and NaV1.5 (gray) with (Ca2+)4-CaM. Simulations of (Ca2+)4-CaM binding the FGF12A LTP and each FGF CaMBD are based on the affinities reported in Table 1. Simulations of (Ca2+)4-CaM binding to IQ motifs are based on Kd of 19.8 nM for NaV1.2 and 92.3 nM for NaV1.5 (data not shown). The yellow shaded region highlights the estimated concentration range of free CaM (90) within the cell. The inset shows the ΔG of (Ca2+)4-CaM binding to the putative A-type FGF CaMBDs (FGF11A/orange, FGF12A/purple, FGF13A/red, and FGF14/blue), FGF12A LPTD (green) and the NaV1.2 IQ motif (black) and NaV1.5 IQ motif (gray). The affinities are arranged from tightest (left) to weakest (right). E, schematic depiction of different models of partially ((Ca2+)2-CaMN, apo CaMC) and fully Ca2+-saturated CaM (CaMN/blue, CaMC/red) binding to the IQ motif (green cylinder) in NaV1.2CTD (green) and/or FGF12A CaMBD (limon cylinder) in full-length FGF12A (purple). The FGF12A LTP is shown as a green cylinder and Ca2+ is shown as yellow circles.