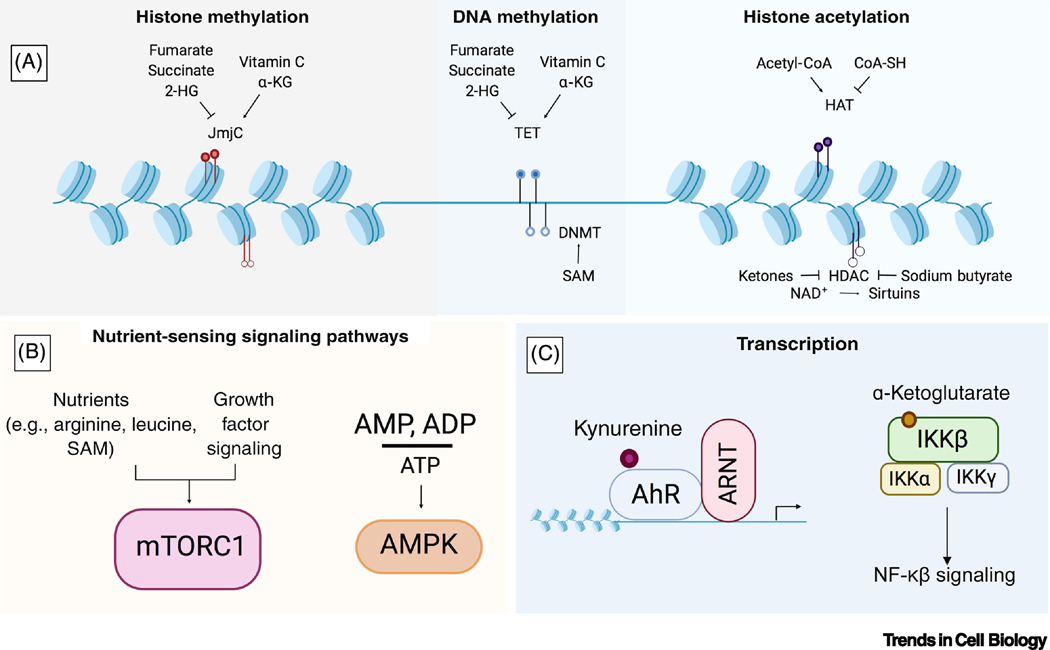
Figure 1. Metabolic Effectors of Chromatin, Signaling Pathways, and Transcription Factors.
(A) Many substrates and co-factors for chromatin-modifying enzymes are derived from metabolic pathways involving the TCA cycle, methionine cycle, folate cycle, glycolysis, β-oxidation, and the hexosamine pathway. These metabolites can serve as activators or inhibitors of epigenetic writers such as Jumonji C (JmjC) domain-containing proteins, DNA methyltransferases (DNMTs), and histone acetyltransferases (HATs), Ten-eleven translocase DNA demethylases (TETs), and histone deacetylases (HDACs). (B) Metabolites can influence nutrient sensing signaling pathways. The kinase complex mTORC1 can only be activated by growth factor-induced signaling when the amino acids arginine and leucine, as well as the co-factor S-adenosyl methionine (SAM), are sensed inside the cell. In addition, energy balance communicated through the cellular AMP/ADP to ATP ratio can be sensed by AMPK. (C) Transcription factors can be directly regulated by metabolites; for example, the tryptophan metabolite kynurenine is an endogenous agonist for the aryl hydrocarbon receptor, and α-KG binds to and activates IKKβ and initiates NF-κβ signaling.