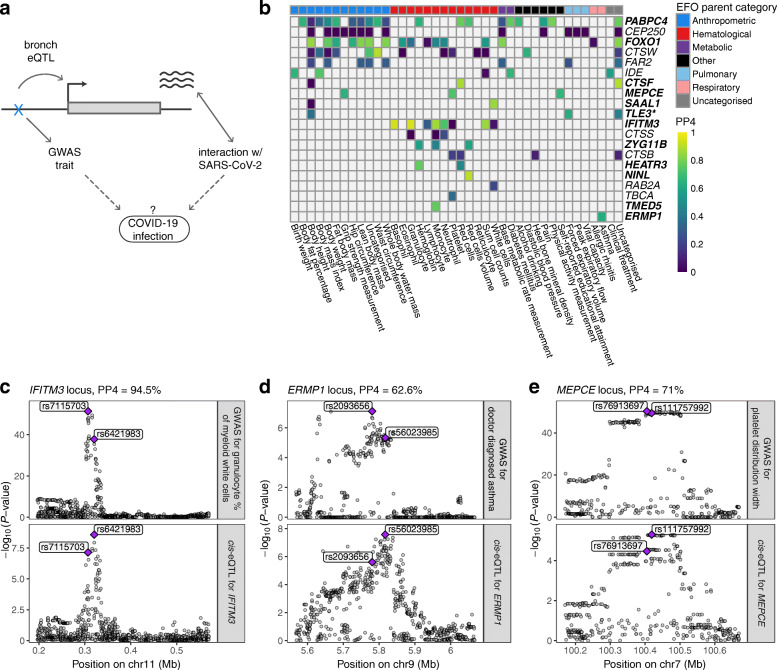
Fig. 5.
Colocalization analysis of the regulatory variants for COVID-19-related genes. a Illustration of the concept of how regulatory variants for COVID-19-related genes in bronchial epithelium can be possible candidates for genetic factors that affect infection or progression of the disease. Dotted lines denote the hypothesis we are able to create by searching for the phenotypic associations of the cis-eQTLs for COVID-19-related genes. b Heatmap of the colocalization analysis results for 20 COVID-19-related genes with eQTLs that have at least one phenotypic association belonging to the experimental factor ontology (EFO) parent categories relevant to COVID-19 (respiratory disease, hematological or pulmonary function measurement). Genes highlighted in bold indicate the loci involving COVID-19-relevant EFO categories with posterior probability for colocalization (PP4) > 0.5, suggesting evidence for shared genetic causality between eQTL and GWAS trait. In the TLE locus, the nearest genome-wide significant variant for forced expiratory volume in 1 s (FEV1) from Shrine et al. [57] is more than 1 Mb away, indicating that the association between the variant and FEV1 might be confounded by incomplete adjustment for height. c–e Regional association plot for the GWAS signal on the upper panel and cis-eQTL signal on the lower panel for IFITM3 (c), ERMP1 (d), and MEPCE (e) locus, where the eQTL for the corresponding gene colocalizes with the GWAS trait relevant to COVID-19. Genomic position of the variants is shown on the x-axis and -log10(P value) of the GWAS or eQTL association on the y-axis. The lead GWAS and eQTL variants are highlighted