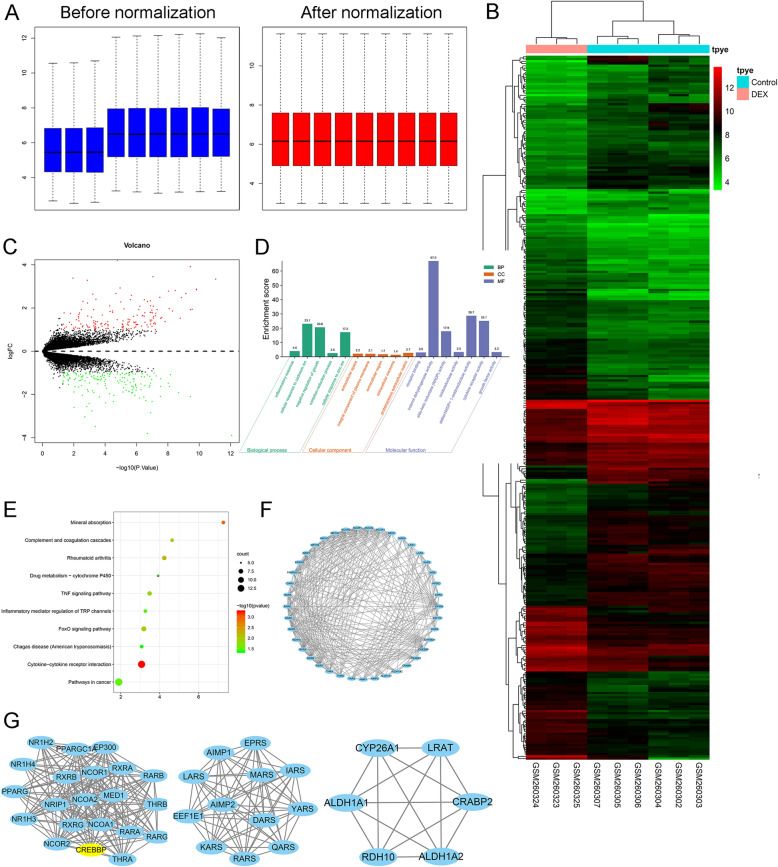
Fig. 1.
Identification of DEGs in human osteoblasts exposed to DEX. a Comparison of expression value between before normalization and after normalization. b Heatmap of expression of differentially-expressed genes: DEX vs. control. c Volcano plot of differentially expressed genes between DEX and control groups. The x-axis represents the -log10 (P value), and the y-axis represents the logFC. d Gene ontology of the differentially expressed genes. The x-axis represents the enriched gene ontology terms, and the y-axis represents the enrichment score. e Top ten KEGG terms for the differentially expressed mRNA. The abscissa represents the enriched score, and the ordinate represents the KEGG pathway terms. P values indicate larger values from blue through to red, and a larger node size represents a higher number of enriched genes. f protein-protein interaction of the differentially expressed genes. g Identification of a top 3 sub-network using MCODE in Cytoscape software