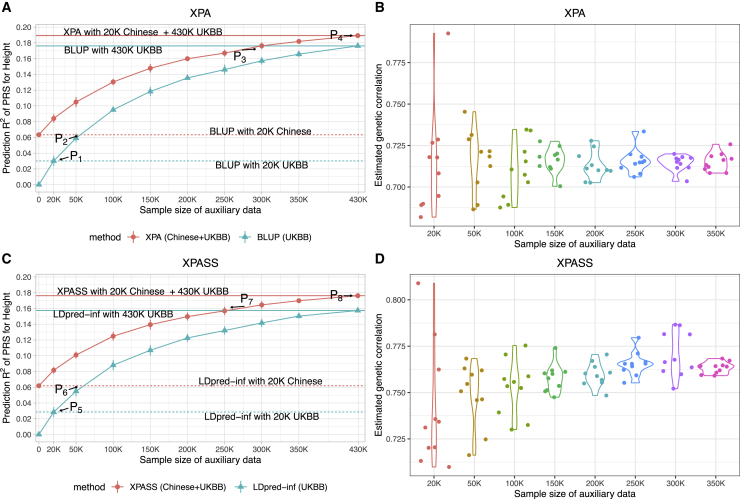
Figure 5.
Influence of the auxiliary sample size on the prediction performance of XPA and XPASS for predicting height
Predictive R2 of XPA and XPASS are shown in (A) and (C). The corresponding trans-ancestry genetic correlations estimated by XPA and XPASS in each replicate are shown in (B) and (D). We trained XPA and XPASS by integrating 21,069 Chinese training samples with 20,000–300,000 random subsamples drawn from UKBB, where samples from UKBB could be viewed as the auxiliary dataset. The results are summarized from ten replications. Dashed horizontal lines in (A) and (C) mark the BLUP/LDpred-inf results obtained by using 20,000 samples from Chinese (red) and UKBB (cyan). Solid horizontal lines in (A) and (C) mark the results obtained by using all UKBB samples with (red) or without (cyan) Chinese. Points P1−P4 in (A) represent the situations where the auxiliary sample size achieves 20,000 (P1), BLUP trained on about 50,000 UKBB samples achieves equivalent performance with that trained on 20,000 Chinese samples (P2), XPA achieves identical performance with BLUP trained on all UKBB samples (P3), and XPA is trained with all UKBB samples (P4). Points P5−P8 in (C) represent the similar situations for summary-level approaches XPASS and LDpred-inf. Error bars represent ±1.96 of the standard error.