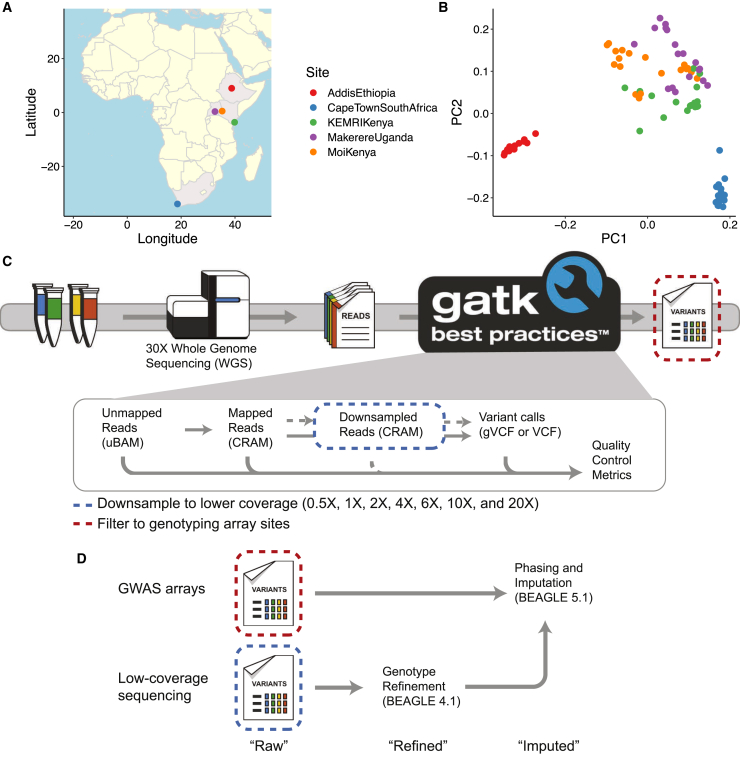
Figure 1.
Populations and sites included in high-coverage whole-genome sequence data and downsampling schema to assess the performance of lower-coverage sequencing versus GWAS arrays
(A) Map indicating where participants in the NeuroGAP-Psychosis study are enrolled in this dataset.
(B) The first two principal components (PCs) show variation within and among populations. They first distinguish the Ethiopians, and then the South Africans, from other African populations. Colors are consistent in (A) and (B).
(C) High-coverage genomes were processed with the GATK best practices pipeline. To mimic lower-coverage sequencing data, we downsampled analysis-ready CRAM files to various depths, followed by a standard implementation of the variant calling pipeline. To mimic GWAS array data, we filtered the variants called from the high-coverage sequencing data to only those sites on the arrays.
(D) After variants were filtered from high-coverage data to sites on GWAS arrays, they were phased and imputed with Beagle 5.1. After downsampling reads from high-coverage data to various depths of coverage, we refined genotypes by using Beagle 4.1 (the last version of Beagle to provide this feature), then phased and imputed them by using Beagle 5.1, as with GWAS arrays. “Raw” indicates that variant calls were produced directly from GATK with no genotype refinement or imputation, “refined” indicates variant calls from genotype refinement without imputation, and “imputed” indicates imputed variants following genotype refinement.