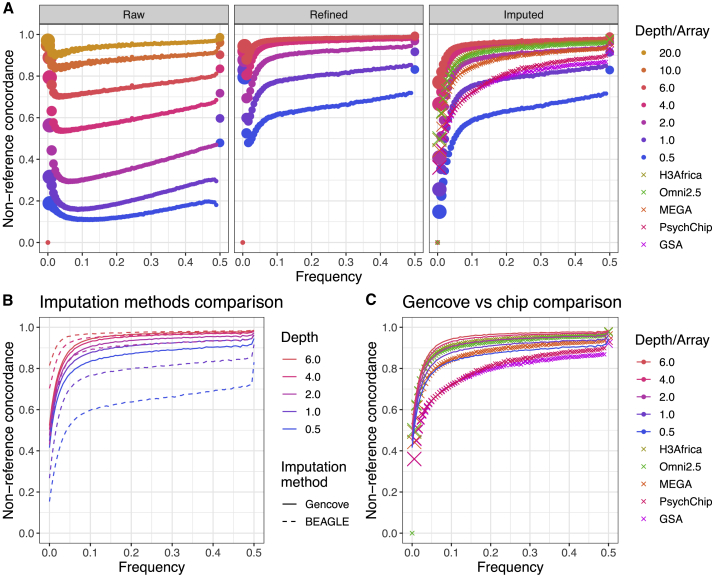
Figure 4.
Non-reference concordance for SNPs as a function of sequencing depth or genotyping array, frequency, analysis stage, and imputation method
“Truth” dataset here is the full depth joint called sequencing dataset. All depths of sequencing data are shown for the raw data (i.e., only variant calling from GATK with no genotype refinement or imputation following). We excluded sequencing at 10× and 20× for all except the raw data because of minimal potential accuracy gains and to reduce computational costs.
(A) Non-reference concordance comparisons throughout steps of the Beagle analysis pipeline. Size of the points are proportional to the number of SNPs in each frequency bin. “Raw” indicates that variant calls were produced directly from GATK with no genotype refinement or imputation, “refined” indicates variant calls from genotype refinement without imputation, and “imputed” indicates imputed variants following genotype refinement.
(B) Non-reference concordance comparisons of Beagle versus Gencove software for imputation of low-coverage data.
(C) Non-reference concordance comparison of Gencove software for imputation of low-coverage data versus Beagle for imputation of GWAS arrays. Non-reference concordance values averaged across (B) and (C) are shown in Table S4.