Summary
The DNA damage-binding protein 1 (DDB1) is part of the CUL4–DDB1 ubiquitin E3 ligase complex (CRL4), which is essential for DNA repair, chromatin remodeling, DNA replication, and signal transduction. Loss-of-function variants in genes encoding the complex components CUL4 and PHIP have been reported to cause syndromic intellectual disability with hypotonia and obesity, but no phenotype has been reported in association with DDB1 variants. Here, we report eight unrelated individuals, identified through Matchmaker Exchange, with de novo monoallelic variants in DDB1, including one recurrent variant in four individuals. The affected individuals have a consistent phenotype of hypotonia, mild to moderate intellectual disability, and similar facies, including horizontal or slightly bowed eyebrows, deep-set eyes, full cheeks, a short nose, and large, fleshy and forward-facing earlobes, demonstrated in the composite face generated from the cohort. Digital anomalies, including brachydactyly and syndactyly, were common. Three older individuals have obesity. We show that cells derived from affected individuals have altered DDB1 function resulting in abnormal DNA damage signatures and histone methylation following UV-induced DNA damage. Overall, our study adds to the growing family of neurodevelopmental phenotypes mediated by disruption of the CRL4 ubiquitin ligase pathway and begins to delineate the phenotypic and molecular effects of DDB1 misregulation.
Keywords: DDB1, intellectual disability, CRL4, mutation
Main text
DNA integrity is essential for proper DNA function and, thus, human health. Preserving DNA integrity is difficult because many cell-intrinsic and cell-extrinsic factors cause thousands of damage events every day.1 The DNA damage response signaling pathway maintains genome stability by sensing damage events and activating relevant DNA repair mechanisms.2 DDB1 (MIM: 60045) encodes the damage-specific DNA-binding protein 1, DDB1, which plays a vital role in the DNA damage response, specifically in the nucleotide excision repair pathway where it functions as part of the CUL4–DDB1 ubiquitin E3 ligase complex (CRL4).3 The CRL4 complex has also been found to function in other cellular processes, including regulation of chromatin remodeling, DNA replication, and signal transduction.4, 5, 6, 7, 8 Haploinsufficiency of CUL4B (MIM: 300304) and PHIP (MIM: 612870), two additional CRL4 complex components, has been shown to lead to overlapping forms of syndromic intellectual disability. Pathogenic variants in CUL4B cause X-linked Cabezas syndrome (MIM: 300354), characterized by intellectual disability, seizures, brain malformations, behavioral issues, central obesity, macrocephaly, hypogonadotropic hypogonadism, and dysmorphic facial features.9 Pathogenic PHIP variants cause Chung-Jansen syndrome (MIM: 617991), characterized by intellectual disability, hypotonia, behavioral issues, obesity, and dysmorphic facial features.10,11
Pathogenic DDB1 variants have not yet been described in humans, although homologs of DDB1 have been shown as essential proteins in model organisms where tissue-specific and global absence of the protein in mice and zebrafish often results in embryonic lethality, but no gross phenotype was observed for heterozygous knockout mice within the first year of life.12, 13, 14
Therefore, it is likely that DDB1 is also an essential gene, and pathogenic variants may cause a human phenotype resembling those of CUL4B- and PHIP-related conditions.
Herein, we present eight unrelated individuals (Figure 1) with de novo heterozygous DDB1 variants. Because each of the affected individuals was an isolated case in their family, they were predicted to have either a de novo or recessive condition and exome/genome sequencing was performed at their respective genetics centers. Assessment of all potential recessive or de novo variants for each affected individual was conducted and a list of candidate variants for each individual is provided in Table S2. Each individual harbored a de novo monoallelic DDB1 variant, each of which was predicted to be likely deleterious (Table S3).
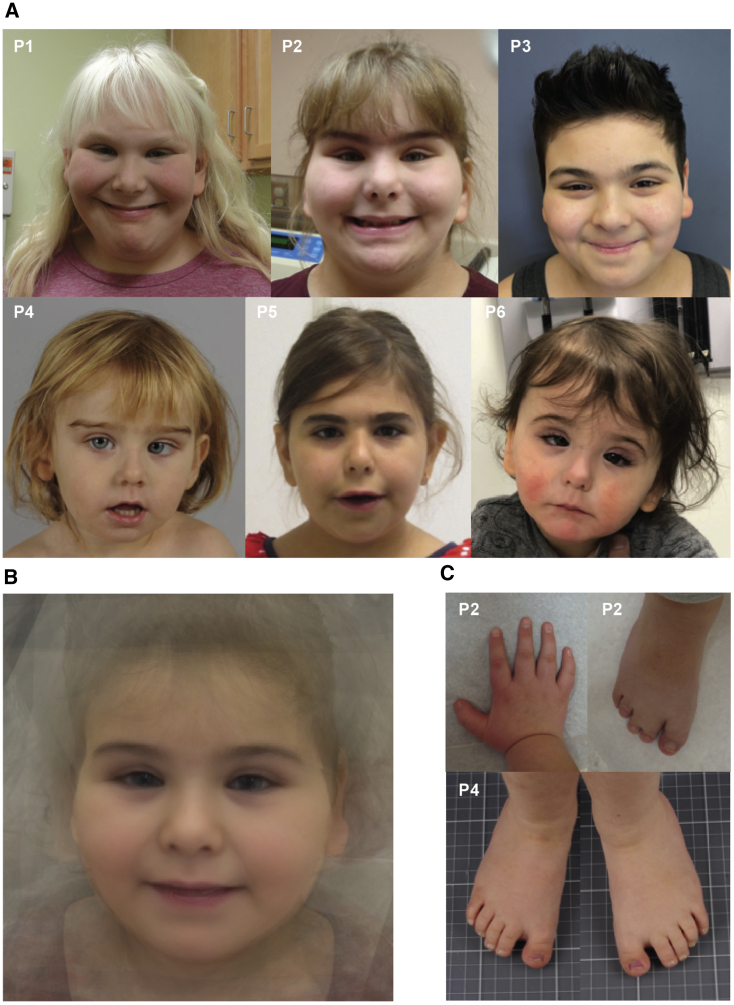
Figure 1.
Face, hand, and feet and composite photographs
(A) Facial photographs of individuals 1, 2, 3, 4, and 5 showing horizontal eyebrows, short nose, and full cheeks.
(B) Composite image of individuals with DDB1 variants showing key distinctive features of horizontal eyebrows, short nose, full cheeks, and large ear lobes.
(C) Photograph of hand in individual 2 showing brachydactyly and proximally placed thumb. Photograph of feet in individuals 2 and 4 showing syndactyly and brachydactyly.
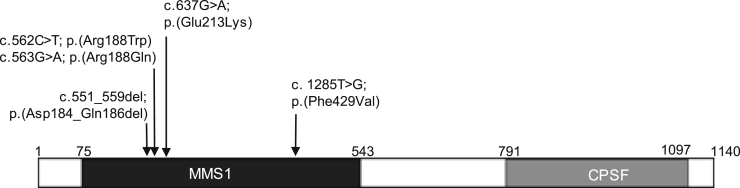
All variants clustered within the first mono-functional DNA-alkylating methyl methanesulfonate (MMS1) domain of DDB1 (Figure 2), and interestingly, we observed one recurrent protein change in four individuals, c.637G>A (GenBank: NM_001923.4) (p.Glu213Lys [GenBank: NP_001914.3]), and two different substitutions at the same residue, p.Arg188Trp and p.Arg188Gln. Each individual’s data were submitted to Matchmaker Exchange with the DDB1 variant’s having been identified as a plausible disease candidate. Detailed phenotype comparison showed a strong clinical overlap, and this cohort of affected individuals was assembled (Table 1). Consistent features were hypotonia (7/8 individuals) and mild-moderate developmental delay or intellectual disability (8/8 individuals). Growth was variable. Two individuals had weight over the 97th centile and all three older individuals had a BMI in the obese range for their age. Malformations occurred in six individuals and included renal (2/8), cardiac (1/8), and anorectal malformations (2/8) and craniosynostosis (1/8). Brachydactyly was common and most noticeable in the feet (6/8 individuals), and two individuals had cutaneous toe syndactyly. Using photographs from the cohort, we generated a composite face.15 This illustrated the key dysmorphic features: horizontal or slightly bowed eyebrows, which were dark in two individuals with fair scalp hair; mild narrowing of the palpebral fissures; full cheeks; a short nose; and large, fleshy and forward-facing ear lobes (Figure 1; see supplemental notes and Table S4 for more detailed phenotypic information).
Figure 2.
Protein domains of DDB1 and de novo variants found in affected individuals
DDB1 contains the MMS1 and CPSF protein domains. The MMS1 domain is homologous to the N-terminal region of MMS1; the protein itself protects against replication-dependent DNA damage in Saccharomyces cerevisiae and belongs to the DDB1 family of CUL4 adaptors. The function of the CPSF domain, homologous to the C terminus of the CPSF A subunit, is unknown but may be involved in RNA/DNA binding. All eight individuals had one de novo DDB1 missense variant, all of which were located within the coding region of the MMS1 domain and are listed above the schematic. Numbers above the schematic denote amino acid positions.
Table 1.
Phenotype information for individuals with DDB1 variants
| Individual | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 |
|---|---|---|---|---|---|---|---|---|
| Gender | female | female | male | female | female | male | male | female |
| Age at last assessment | 17 y | 9 y 2 m | 10 y 8 m | 3 y | 13 y | 22 m | 2 y 11 m | 1 y |
| Molecular data | ||||||||
| DDB1 variant (g) Hg19 | chr11: 61094361_61094369del | chr11: 61094353G>A | chr11: 61094352C>T | chr11: 61094278C>T | chr11: 61083980A>C | |||
| DDB1 variant (c) (NM_001923.4) variant (p) (NP_001914.3) | c.551_559del (p.Asp184_Gln186del) | c.562C>T (p.Arg188Trp) | c.563G>A (p.Arg188Gln) | c.637G>A (p.Glu213Lys) | c.1285T>G (p.Phe429Val) | |||
| Phenotypic features | ||||||||
| Hypotonia | moderate | mild | nil | moderate | moderate | moderate | moderate | moderate |
| Intellectual disability | moderate | moderate DD | mild (IQ = 69) | mild | mild-moderate | mild | moderate DD | moderate DD |
| Craniofacial | thick, light-blonde eyebrows; deep-set, upslanting palpebral fissures; epicanthic folds; short nose in early childhood; very full cheeks; thin upper vermilion; wide mouth; large ears with large and long ear lobes | horizontal, dark eyebrows synophrys; long palpebral fissures with lateral extension; epicanthus inversus; long dark eyelashes; short nose; underdeveloped alae nasi; mid-face hypoplasia; full cheeks; low-set, large ears with long fleshy lobes | synophrys; short palpebral fissures; full cheeks; large earlobes | horizontal, dark eyebrows with medial broadening; lateral extension; lateral extension to palpebral fissures; short nose; mid-face hypoplasia; thin upper vermilion; large ears with fleshy lobes | dark, horizontal, heavy eyebrows; round face; mild midface hypoplasia; full cheeks; protruding upper lip; thin vermillion retrognathia asymmetric; occlusion of teeth; large fleshy ears and earlobes | medial broadening of eyebrows; telecanthus; lateral extension to palpebral fissures; epicanthus; long eyelashes; short nose, retrognathia; thin, tented upper vermilion; low set, fleshy ears | deep-set eyes; epicanthus; hypotelorismconvergent strabismus; nystagmus; short and upturned nose; underdeveloped alae nasi; full cheeks; large, simple ears | medial broadening of eyebrows; telecanthus proptosis epicanthus inversus; flat nasal bridge; mid-face hypoplasia |
| Malformations | anterior anus and recto-vaginal fistula | accessory band across left ventricle of heart, horseshoe kidney, anterior ectopic anus, dysgenesis of corpus callosum | nil | nil | horseshoe kidney with left vesico-ureteric reflux, pelvicalyceal dilatation and megaureter | nil | mild left hydronephrosis | metopic cranio-synostosis |
| Hands and feet | small hands and feet, short fourth metacarpals, brachydactyly | 2–3 toe syndactyly, brachydactyly | short toes | short toes | short toes | NR | 2–3 toe partial syndactyly | NR |
| Other | joint laxity, ADHD, anxiety, hypothyroidism, recurrent otitis media, obesity | joint laxity, truncal obesity | frequent otitis media | mild joint laxity, bilateral hip dysplasia, obstructive sleep apnea | mild joint laxity | NR | gastro-esophageal reflux | NR |
NR, not reported; DD, developmental delay.
An assessment of gnomAD showed that DDB1 is missense- and CNV-depleted (Z = 5.67 and 1.30, respectively) and intolerant to LoF variants (pLI = 1.0, o/e = 0.13 [0.08–0.24]). Eukaryotic homologs of DDB1 were aligned, and conservation among the sequences is quite high, notably including the residues of interest in the affected individuals, which further demonstrates conservation of these sites (Figure S1C). Given the highly similar phenotype observed in these individuals with de novo apparently deleterious variants in what appears to be an essential protein, DDB1 represented a strong candidate gene for the observed phenotypes.
All DDB1 variants were missense, excepting the deletion found in individual 1. To assess the impact of the DNA deletion at the intron 4-exon 5 boundary found in individual 1, we performed splicing studies by using lymphoblasts from individual 1. This revealed a deletion of nucleotides c.551_559 within the transcript, corresponding to p.Asp184_Gln186del, and immunoblot analysis demonstrated no noticeable change in protein levels as a result (Figures S1A and S1B). On the basis of this result, it is not clear whether this variant is a gain- or loss-of-function variant, similar to the other variants in the cohort.
As our cohort’s variants consisted of several missense variants and one in-frame deletion in the MMS1 domain of DDB1, the mutation mechanism was unclear. To determine whether DDB1 was sensitive to dosage perturbation and associated with human phenotypes when deleted or duplicated, we interrogated the Decipher database for deletions or duplications of DDB1.16 No individuals were identified where haploinsufficiency for DDB1 was associated with a phenotype. One individual was identified with a maternally inherited duplication including the DDB1 gene and another individual had a de novo 134 Mb deletion including DDB1 along with many other genes. No further conclusions could be garnered from these observations.
We next sought to investigate DDB1 function in lymphoblasts from individuals 2 and 4. A key function of DDB1 is to recognize and bind to areas of UV-induced DNA damage. We began by evaluating DDB1 levels in lymphoblast cells from control and affected individuals under basal conditions, as well as following UV induction of DNA damage. All experiments were conducted with a minimum of three replicates with different controls, and representative images are shown. Immunoblot analysis of total protein extracts and qPCR of mRNA showed somewhat variable levels of DDB1 before and after DNA damage, all of which were within the range observed in controls (Figures 3A, 3B, and S2A). We conclude that overall DDB1 mRNA and protein levels are not significantly altered in cells from affected individuals. On the basis of these analyses, however, we cannot distinguish whether mutant DDB1 is expressed equivalently to wild-type or whether there is compensation from the opposing allele.
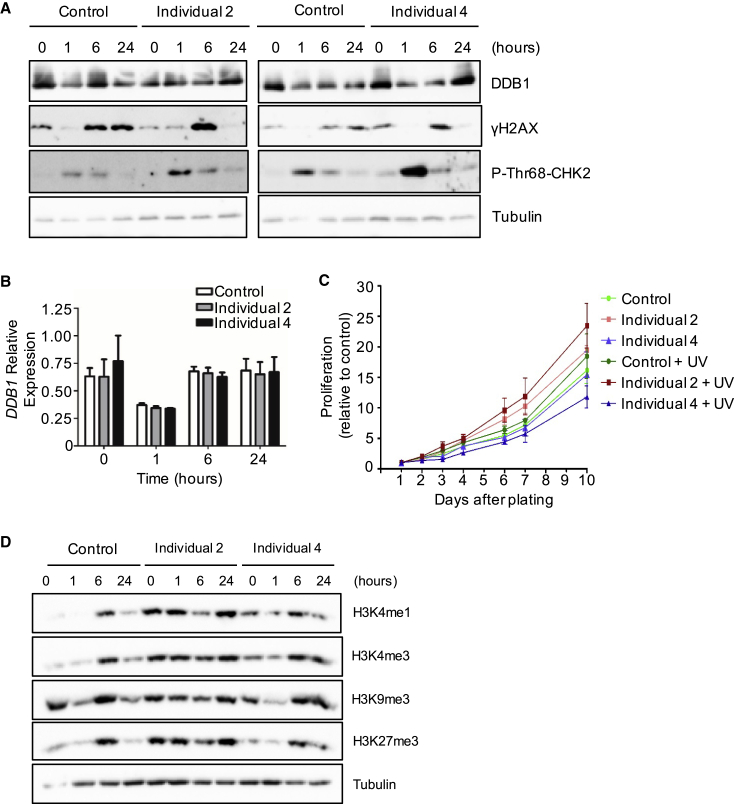
Figure 3.
DDB1 missense variants in lymphoblast cells result in altered DNA damage signatures and histone methylation following UV damage
(A) Immunoblot analysis on total extracts from control and affected lymphoblast cells. Untreated cells are shown at the 0 h time point, whereas the other time intervals indicate the number of h following UV exposure. Total DDB1 and the levels of γH2AX and p-Thr68-CHK2 phosphorylation were assessed: DDB1 was found to be unchanged and γH2AX and p-Thr68-CHK2 levels were induced as expected, p-Thr68-CHK2 to a higher level than controls and γH2AX to a similar level as controls but was not sustained.
(B) Real-time PCR analysis on extracts from lymphoblast cells showing transcript levels of DDB1 before and after UV exposure are similar between cells from affected individuals and control individuals.
(C) Cell proliferation of control and affected lymphoblast cells was measured by harvesting and counting cells on each of the specified days after initial plating, either with or without UV exposure.
(D) Immunoblot analysis of total extracts from control and affected lymphoblast cells. Untreated cells are shown at the 0 h time point, whereas the other time intervals indicate the number of h following UV exposure. Levels of various histone H3 methylations were assessed and found to be abnormal in cells from affected individuals. Immunoblots in this figure are representative images of at least three biological replicates, and graphed data represent the mean of three biological replicates; error bars depict standard error of the mean.
To assess DNA damage signaling we began by measuring γH2AX, a marker of DNA damage that localizes to double-stranded breaks. Cells were again treated with UV, and immunoblot analysis was conducted. We found that γH2AX is induced in both control and affected lymphoblasts, as expected, but these levels are not sustained for as long after DNA damage in affected cells compared to controls. We next assessed a second marker of DNA damage response, phosphorylation of Thr68 on CHK2 (p-Thr68-CHK2), a protein kinase that functions in the cell cycle arrest and apoptosis pathways following DNA damage. As expected, p-Thr68-CHK2 was increased after UV exposure in all cells, but interestingly, these levels appear noticeably higher 1 h after DNA damage in both affected samples compared to control lymphoblasts (Figures 3A and S2A). These data suggest that the DNA damage response is altered in DDB1 cells from affected individuals. We next sought to investigate whether an altered DNA damage response affected growth of these cells. Cellular proliferation of the lymphoblast cells was assessed through cell counts at days 0–10, and we determined that the DDB1 variants do not cause altered lymphoblast proliferation rates compared to control (Figures 3C and S2B). Moreover, when cells were subjected to DNA damage by UV, despite differences in DNA damage signaling, no significant differences in proliferation were observed between control and affected lymphoblasts (Figures 3C and S2B).
Given the altered DNA damage signaling in cells from affected individuals, we hypothesized that other DDB1-regulated pathways may also be affected. The CUL4-DDB1 complexes interact with multiple WD40-repeat proteins, including WDR5 and the Polycomb-group protein EED, which are core components of histone methylation complexes necessary for histone H3 methylation at K4 or K27, respectively.4,17,18 More recently, it has also been shown that DDB1- and CUL4-associated factor 8 (DCAF8), another WD40-repeat protein, interacts with CRL4 to ubiquitinate H3 in adult mouse hepatocytes, which promotes methylation of H3K9.19 Because of these associations between H3 methylation and WD40-repeat proteins that interact with CRL4, we investigated whether there were any changes in H3K4, H3K9, and H3K27 methylation in cells from affected individuals. H3K27me3 did not seem to be affected by DDB1 variants, as levels were increased at 6 h and 24 h after UV exposure in both control and lymphoblast cells from affected individuals (Figures 3D and S2C). The general trend for H3K4me1, H3K4me3, and H3K9me3 was increased levels at 6 h after DNA damage in control lymphoblasts, which start to return to basal levels after 24 h (Figures 3D and S2C). In both affected lymphoblast cells, H3K4me1 and H3K4me3 protein levels appeared to be increased compared to control cells without induction of DNA damage. H3K4 and H3K9 methylation in affected lymphoblasts were then increased later than in the control after UV exposure, sometimes beginning 6 h after UV and generally elevated 24 h after treatment as well. However, the increase in these histone modifications in lymphoblasts from affected individuals following DNA damage was sometimes subtle because of the abundance of these marks at baseline. Together, these results suggest that increased histone methylation may occur later and may also be sustained for longer periods of time following UV exposure in cells from affected individuals (Figure 3D). Given the abnormal DDB1 downstream effects, and the identification of eight individuals with a highly similar phenotype, we conclude that we have identified a unique condition caused by monoallelic de novo variants in the MMS1 domain of DDB1.
DDB1 functions in a complex with CUL4 and PHIP. Loss-of-function variants in genes encoding these proteins, CUL4B and PHIP, have been observed in individuals with syndromic intellectual disability. This suggests a common pathogenic mechanism for this family of phenotypes.9,10 Seven of the eight individuals in this have missense variants in DDB1, including one recurrent protein change, p.Glu213Lys, and two different substitutions at the same amino acid residue (p.Arg188Trp and p.Arg188Gln), while the remaining variant is an in-frame deletion of three amino acids. This suggests these may function through a dominant negative or gain-of-function mechanism, although loss-of-function cannot be excluded.
Data from a recent study investigating regional missense variant constraint demonstrate that the DDB1 transcript has two distinct sub-genic regions, both of which are observed to contain less than half of the expected number of missense variants and together span the entire gene.20 Interestingly, transcripts and regions of transcripts that have the fraction of expected variation observed as ≤0.6 represent 14% of the coding region of the human genome but contain 89% of all the ClinVar pathogenic missense variants.20 Therefore, it remains a possibility that because the entire DDB1 transcript is depleted of missense variants, variants outside of the MMS1 domain may still have yet to be identified as contributing to this neurodevelopmental disorder. This is further supported by the fact that the majority of DDB1 is comprised of three WD40 β-propeller structures; the affected residues Asp184 to Glu213 are all present in one of them, Phe429 is present in the second, while the third remaining WD40 β-propeller overlaps the CPSF domain.21 Because WD40 β-propellers have no catalytic activity and instead function together as protein-protein or protein-DNA interaction platforms, alterations within these regions most likely impact a number of overlapping DDB1 interactions.22 Although the functional studies showed clear effects downstream of DDB1, it was not possible to discern a mutational mechanism. For the variants p.Asp184_Gln186del, p.Arg188Gln, and p.Phe429Val, present in individuals 1, 3, and 8, respectively, no functional studies were able to be undertaken because cells were not available. Nevertheless, the phenotypic similarity across the cohort led us to conclude that the rare, deleterious DDB1 variants in individuals 1, 3, and 8 were also highly likely to be pathogenic. Phenotypic features in common across the CUL4B-, PHIP-, and DDB1-related conditions include intellectual disability and obesity, and there is some intriguing similarity in the facial dysmorphism of all three phenotypes and digital features in the CUL4B- and DDB1-related phenotypes.9, 10, 11 In contrast to our cohort, brain malformations, tremor, macrocephaly, and hypogonadotropic hypogonadism are common features in Cabezas syndrome and behavioral issues are frequently reported for both Cabezas and Chung-Jansen syndromes.
The common phenotype of obesity is interesting because there is growing evidence linking the CRL4 complex with control of adipogenesis. WDTC1, which is known to suppress adipogenesis, is a member of the CRL4 complex. Obesity is observed in humans and mice when expression of WDTC1 is reduced.23, 24, 25 Moreover, disruption of the interaction between WDTC1 and DDB1 in vitro leads to adipogenic gene expression.26 The CRL4 complex also has a role in the regulation of glucose metabolism, promoting hepatic gluconeogenesis.27 Of interest, one individual in our study had hypoglycemia in the neonatal period, and it is therefore possible that the DDB1 variant in this individual adversely affected the control of glucose homeostasis.
DDB1 and DDB2 form the UV-DDB complex, which is integral to the nucleotide excision repair process.3 Bi-allelic variants in DDB2 (MIM: 600811) cause xeroderma pigmentosum group E (MIM: 278740), a neurocutaneous syndrome causing skin cancers and dermatological and ocular features.28 Although DDB1 is a protein involved in DNA repair, we did not observe the cardinal phenotypes seen in DNA repair disorders, such as growth restriction, skin alterations, premature aging, and a predisposition to malignancy. In addition, most DNA repair disorders are caused by bi-allelic loss-of-function variants. These findings further support a dominant negative or gain-of-function mechanism for the DDB1 variants in this study or could be explained by an additional yet unknown function for DDB1. In lymphoblasts from affected individuals from our study, we find altered DNA damage signaling through changes in γH2AX and Chk2 phosphorylation, consistent with DDB1’s playing an important role in repair following UV damage. Somatic DDB1 variants are reported in breast, lung, and gastrointestinal malignancies, and the CRL4 complex demonstrates aberrant function in many different malignancies.29 Interestingly, within the Catalogue Of Somatic Mutations In Cancer (COSMIC), missense variants at the Arg188 residue are found in nine samples, making it the most frequently affected amino acid of DDB1.29 Of the samples in COSMIC with missense variants at the Arg188 residue, p.Arg188Trp is one of the nine and p.Arg188Gln represents another five, although neither of these variants were studied to determine their functional impact, so it is unclear whether DDB1 acts primarily as a tumor suppressor or oncogene in these contexts. Although there is no cancer involvement in the cohort described here, this may represent yet another gene family in which germline variants cause a neurodevelopmental phenotype and somatic variants cause cancer.29,30
In summary, we have used genomic sequencing to identify de novo variants in DDB1 in eight unrelated individuals with overlapping phenotypes of intellectual disability and hypotonia with a facial gestalt of maxillary hypoplasia, small nose, lateral extension of the palpebral fissures, and straight eyebrows. Functional characterization in cells from affected individuals showed altered DDB1 function resulting in abnormal DNA damage signatures and histone methylation following UV-induced DNA damage. Our findings extend the disease association of components of the CRL4 and UV-DDB complexes, inform the phenotypic understanding of this disorder, and provide insight into the molecular pathogenesis of this condition.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
The authors would like to thank the individuals and their families for their generous participation in this and the many healthcare providers involved in their care. We thank Natalie Tan for her assistance with the photographic figure. Funding for the UDP-Vic was provided by philanthropic donation and the Murdoch Children's Research Institute. The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program, the Harbig Family Foundation, and The Royal Children's Hospital Foundation. This work was supported, in part, by the Care4Rare Canada Consortium funded by Genome Canada and the Ontario Genomics Institute (OGI-147), the Canadian Institutes of Health Research, Ontario Research Fund, Genome Alberta, Genome British Columbia, Genome Quebec, and Children's Hospital of Eastern Ontario Foundation. Sequencing and analysis was provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung, and Blood Institute grant UM1 HG008900 to Daniel MacArthur and Heidi Rehm. Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under award number U01HG009599. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. P.J.L. is supported by the Vincent Chiodo Foundation. D.W. and K.Õ. are members of the European Reference Network ITHACA. K.Õ. and S.P. were supported by Estonian Research Council grants PRG471, MOBTP175, and PUTJD827.
Published: March 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.03.007.
Data and code availability
There are restrictions to the availability of genomic sequencing data in this project because of consent limitations.
Web resources
Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org/
Matchmaker Exchange, https://www.matchmakerexchange.org/
OMIM, https://omim.org/
Supplemental information
References
- 1.Tiwari V., Wilson D.M., 3rd DNA Damage and Associated DNA Repair Defects in Disease and Premature Aging. Am. J. Hum. Genet. 2019;105:237–257. doi: 10.1016/j.ajhg.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maréchal A., Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scrima A., Konícková R., Czyzewski B.K., Kawasaki Y., Jeffrey P.D., Groisman R., Nakatani Y., Iwai S., Pavletich N.P., Thomä N.H. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higa L.A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 5.Nishitani H., Sugimoto N., Roukos V., Nakanishi Y., Saijo M., Obuse C., Tsurimoto T., Nakayama K.I., Nakayama K., Fujita M. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh P., Wu M., Zhang H., Sun H. mTORC1 signaling requires proteasomal function and the involvement of CUL4-DDB1 ubiquitin E3 ligase. Cell Cycle. 2008;7:373–381. doi: 10.4161/cc.7.3.5267. [DOI] [PubMed] [Google Scholar]
- 7.Hrecka K., Gierszewska M., Srivastava S., Kozaczkiewicz L., Swanson S.K., Florens L., Washburn M.P., Skowronski J. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. USA. 2007;104:11778–11783. doi: 10.1073/pnas.0702102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong X., Zhang D., Guha A., Arthurs B., Cazares V., Gupta N., Yin L. CUL4-DDB1-CDT2 E3 Ligase Regulates the Molecular Clock Activity by Promoting Ubiquitination-Dependent Degradation of the Mammalian CRY1. PLoS ONE. 2015;10:e0139725. doi: 10.1371/journal.pone.0139725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vulto-van Silfhout A.T., Nakagawa T., Bahi-Buisson N., Haas S.A., Hu H., Bienek M., Vissers L.E.L.M., Gilissen C., Tzschach A., Busche A. Variants in CUL4B are associated with cerebral malformations. Hum. Mutat. 2015;36:106–117. doi: 10.1002/humu.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webster E., Cho M.T., Alexander N., Desai S., Naidu S., Bekheirnia M.R., Lewis A., Retterer K., Juusola J., Chung W.K. De novo PHIP-predicted deleterious variants are associated with developmental delay, intellectual disability, obesity, and dysmorphic features. Cold Spring Harb. Mol. Case Stud. 2016;2:a001172. doi: 10.1101/mcs.a001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen S., Hoischen A., Coe B.P., Carvill G.L., Van Esch H., Bosch D.G.M., Andersen U.A., Baker C., Bauters M., Bernier R.A. A genotype-first approach identifies an intellectual disability-overweight syndrome caused by PHIP haploinsufficiency. Eur. J. Hum. Genet. 2018;26:54–63. doi: 10.1038/s41431-017-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cang Y., Zhang J., Nicholas S.A., Bastien J., Li B., Zhou P., Goff S.P. Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell. 2006;127:929–940. doi: 10.1016/j.cell.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 13.Cang Y., Zhang J., Nicholas S.A., Kim A.L., Zhou P., Goff S.P. DDB1 is essential for genomic stability in developing epidermis. Proc. Natl. Acad. Sci. USA. 2007;104:2733–2737. doi: 10.1073/pnas.0611311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z., Holzschuh J., Driever W. Loss of DDB1 Leads to Transcriptional p53 Pathway Activation in Proliferating Cells, Cell Cycle Deregulation, and Apoptosis in Zebrafish Embryos. PLoS ONE. 2015;10:e0134299. doi: 10.1371/journal.pone.0134299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reijnders M.R.F., Miller K.A., Alvi M., Goos J.A.C., Lees M.M., de Burca A., Henderson A., Kraus A., Mikat B., de Vries B.B.A., Deciphering Developmental Disorders Study De Novo and Inherited Loss-of-Function Variants in TLK2: Clinical and Genotype-Phenotype Evaluation of a Distinct Neurodevelopmental Disorder. Am. J. Hum. Genet. 2018;102:1195–1203. doi: 10.1016/j.ajhg.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon J.A., Kingston R.E. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 18.Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 19.Li G., Ji T., Chen J., Fu Y., Hou L., Feng Y., Zhang T., Song T., Zhao J., Endo Y. CRL4DCAF8 Ubiquitin Ligase Targets Histone H3K79 and Promotes H3K9 Methylation in the Liver. Cell Rep. 2017;18:1499–1511. doi: 10.1016/j.celrep.2017.01.039. [DOI] [PubMed] [Google Scholar]
- 20.Samocha K.E., Kosmicki J.A., Karczewski K.J., O’Donnell-Luria A.H., Pierce-Hoffman E., MacArthur D.G., Neale B.M., Mark J., Daly M.J. Regional missense constraint improves variant deleteriousness prediction. bioRxiv. 2017 doi: 10.1101/148353. [DOI] [Google Scholar]
- 21.UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C., Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galgani J.E., Kelley D.E., Albu J.B., Krakoff J., Smith S.R., Bray G.A., Ravussin E., Look AHEAD Adipose Research Group Adipose tissue expression of adipose (WDTC1) gene is associated with lower fat mass and enhanced insulin sensitivity in humans. Obesity (Silver Spring) 2013;21:2244–2248. doi: 10.1002/oby.20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai C.-Q., Parnell L.D., Arnett D.K., García-Bailo B., Tsai M.Y., Kabagambe E.K., Straka R.J., Province M.A., An P., Borecki I.B. WDTC1, the ortholog of Drosophila adipose gene, associates with human obesity, modulated by MUFA intake. Obesity (Silver Spring) 2009;17:593–600. doi: 10.1038/oby.2008.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suh J.M., Zeve D., McKay R., Seo J., Salo Z., Li R., Wang M., Graff J.M. Adipose is a conserved dosage-sensitive antiobesity gene. Cell Metab. 2007;6:195–207. doi: 10.1016/j.cmet.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groh B.S., Yan F., Smith M.D., Yu Y., Chen X., Xiong Y. The antiobesity factor WDTC1 suppresses adipogenesis via the CRL4WDTC1 E3 ligase. EMBO Rep. 2016;17:638–647. doi: 10.15252/embr.201540500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong X., Zhang D., Charney N., Jin E., VanDommelen K., Stamper K., Gupta N., Saldate J., Yin L. DDB1-mediated CRY1 degradation promotes FOXO1-driven gluconeogenesis in liver. Diabetes. 2017;66:2571–2582. doi: 10.2337/db16-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang J., Chu G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair (Amst.) 2002;1:601–616. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonell L.M., Kernohan K.D., Boycott K.M., Sawyer S.L. Receptor tyrosine kinase mutations in developmental syndromes and cancer: two sides of the same coin. Hum. Mol. Genet. 2015;24(R1):R60–R66. doi: 10.1093/hmg/ddv254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are restrictions to the availability of genomic sequencing data in this project because of consent limitations.