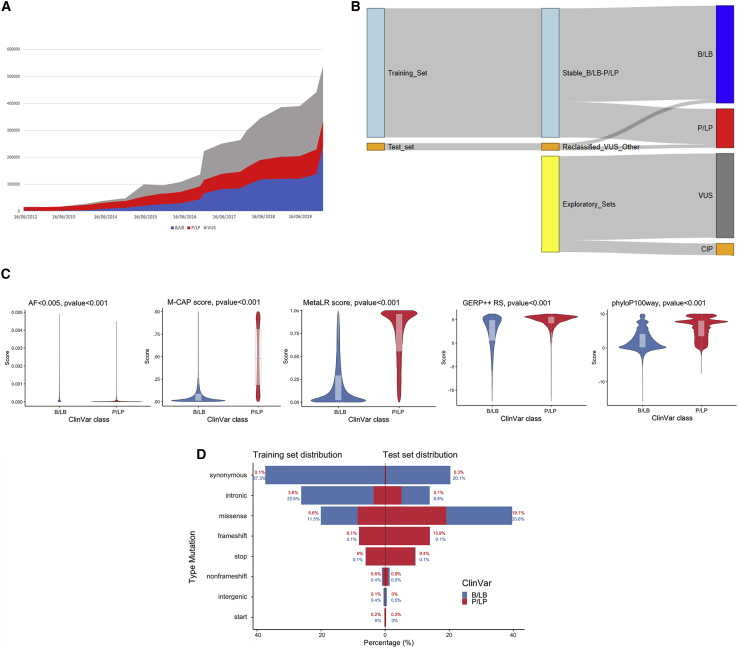
Figure 1.
ClinVar datasets overview
(A) Variants over time. Trend of the total number of variants present in ClinVar, divided by the three main categories of clinical significance: blue for benign (B/LB) variants growth, red for pathogenic (P/LP), and gray for VUSs.
(B) Sankey diagram showing the construction of the different datasets coming from ClinVar. RENOVO training and test set come both from B/LB and P/LP variants that never changed classification and that were reclassified, respectively. VUSs and conflicting interpretation of pathogenicity (CIP) variants are used as an application of RENOVO.
(C) Feature distribution: violin plots for four numerical features of the training set are displayed (AF < 0.005, M-CAP and Meta-LR functional scores, GERP++_RS and phyloP100way_vertebrate conservation scores). Blue is used for distribution in the B/LB class and red for the P/LP class. Boxplots are shown in gray. p values from Wilcoxon rank-sum test are added for each feature.
(D) Variant type distributions in training set (left) and test set (right). For each mutation type, the percentage of B/LB and P/LP variants over the total in the corresponding set is displayed. Blue is used for the B/LB class and red for the P/LP class.