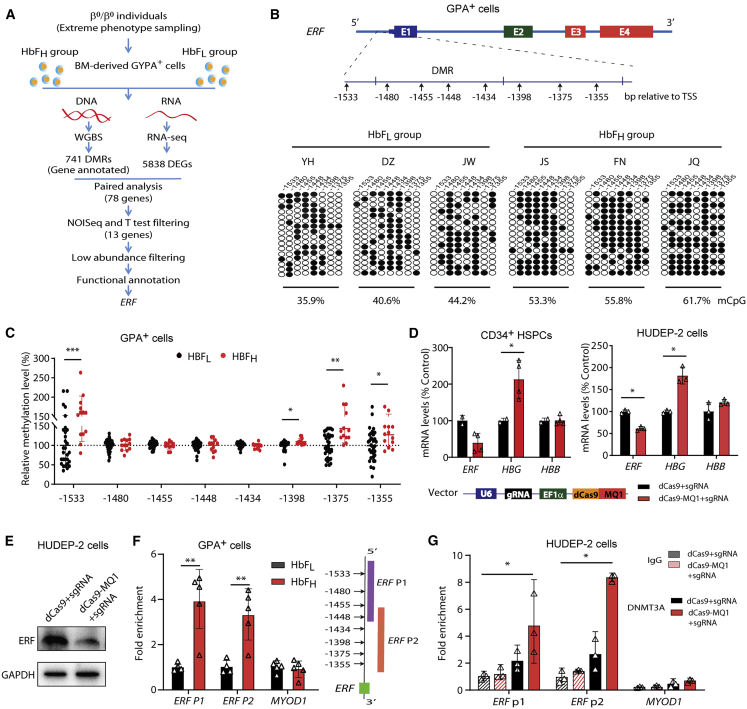
Figure 1.
Identification of ERF as an HbF repressor through transcriptomic and methylation studies in β-thalassemia-affected individuals
(A) A flowchart for the screening of candidate modulators of HbF. The threshold for a positive DMR is defined as differential methylation level ≥ 10% (see the supplemental materials). We screened 741 out of the top 3,000 DMRs (top 3,000 hyper-DMR or hypo-DMR), which are annotated to be located in the promoter regions. 5,835 DEGs were identified according to the following criteria: |log2FC| > 0.8 and divergence probability > 0.8 via NOISeq. 78 genes were enrolled in the candidate list as they presented significantly differential expression in both mRNA and methylation level between HbFL and HbFH groups. We further screened 13 out of the 78 DE-mRNAs (|log2FC| > 0.8 and divergence probability > 0.8 in the NOISeq analysis and p < 0.1 in the further Student’s t test) containing DMRs in their promoter regions, from which we prioritized six candidate genes by filtering out the low abundance with FPKM < 1.0 in either the HbFH or HbFL group. We finally performed functional annotation and identified ERF as the candidate gene with top priority for functional validation.
(B) Top: schematic of ERF gene body. Bottom: analysis of methylation levels at CpG sites (indicated by the distance relative to the transcription start site, TSS) within the DMR in the ERF promoter, evaluated by sequencing. Data were generated with BM-derived GYPA+ cells from individuals with β0-thalassemia (HbFH: n = 3 or HbFL: n = 3) and used in WGBS assays. Each row of eight CpG sites within a group represents a single bisulfite-treated clone with methylated CpGs (•) or unmethylated CpGs (○).
(C) Quantitative measurement of methylation levels of the ERF promoter by bisulfite pyrosequencing in an independent cohort of 47 samples with β-thalassemia (HbFH: n = 13 or HbFL: n = 34). Methylation percentage of each subject relative to the mean level for the subjects with HbFL (the horizontal dashed line) is shown for each CpG site.
(D) Effects of ERF promoter hypermethylation by dCas9-MQ1/sgRNA on ERF, HBG, and HBB expression levels in CD34+ HSPCs (left) and HUDEP-2 cells (right). The schematic of dCas9-MQ1-sgRNA assay is shown in the bottom. MQ1 was methyltransferase that mediated hypermethylation in the target DNA region according to the guide of sgRNA. Two sgRNAs were designed to cover the region of eight CpG sites in the ERF promoter.
(E) Immunoblotting analysis in control and ERF promoter-targeted methylated HUDEP-2 cells. GAPDH served as a loading control.
(F) ChIP-quantitative real-time PCR assays performed with DNMT3A antibody in GYPA+ cells from the HbFH group (n = 5) and HbFL group (n = 5). ERF P1 and P2 covered the region of eight CpG sites in the ERF promoter, as shown in the right panel.
(G) ChIP-quantitative real-time PCR assays of DNMT3A performed in control (dCas9-sgRNA) and methylated (dCas9-MQ1-sgRNA) HUDEP-2 cells. MYOD1 served as the negative control. Data from ≥3 independent experiments are presented as means ± SD (∗p < 0.05; ∗∗p < 0.01).