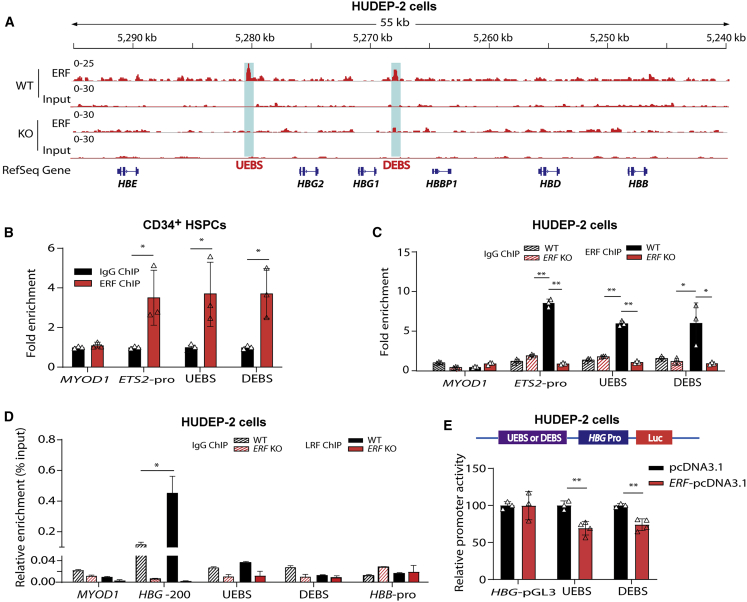
Figure 4.
Identification of ERF-binding sites in β-globin gene clusters
(A) ChIP-seq binding patterns from HUDEP-2 cells with anti-ERF antibody at the β-globin cluster. Two ERF-binding positive signals 3.6 kb upstream of HBG2 (named UEBS) and 1.5 kb downstream of HBG1 (named DEBS) are marked by the light blue shadows.
(B) Detection of ERF-binding sites by ChIP-quantitative real-time PCR in human CD34+ HSPCs.
(C and D) Detection of ERF-binding (C) or LRF-binding (D) activity in ERF KO HUDEP-2 cells (clone #2–6). IgG served as the negative control for ChIP. MYOD1 and HBB-pro served as the negative control for quantitative real-time PCR. ETS2-pro served as the positive control for quantitative real-time PCR. HBG-200, −200 bp upstream of the TSS in the HBG promoter, served as the positive control for LRF binding.
(E) Top: the schematic of dual luciferase reporter assay. Bottom: effects of UEBS or DEBS as regulatory elements on HBG promoter activity in a pGL3 luciferase reporter in HUDEP-2 cells without (black) or with (bright red) ERF overexpression (OE). The data of each group were normalized to the control (pcDNA3.1). Data are presented as means ± SD (∗p < 0.05; ∗∗p < 0.01).