Figure 4.
An overview of the candidate causal variants in autoimmune GWAS loci
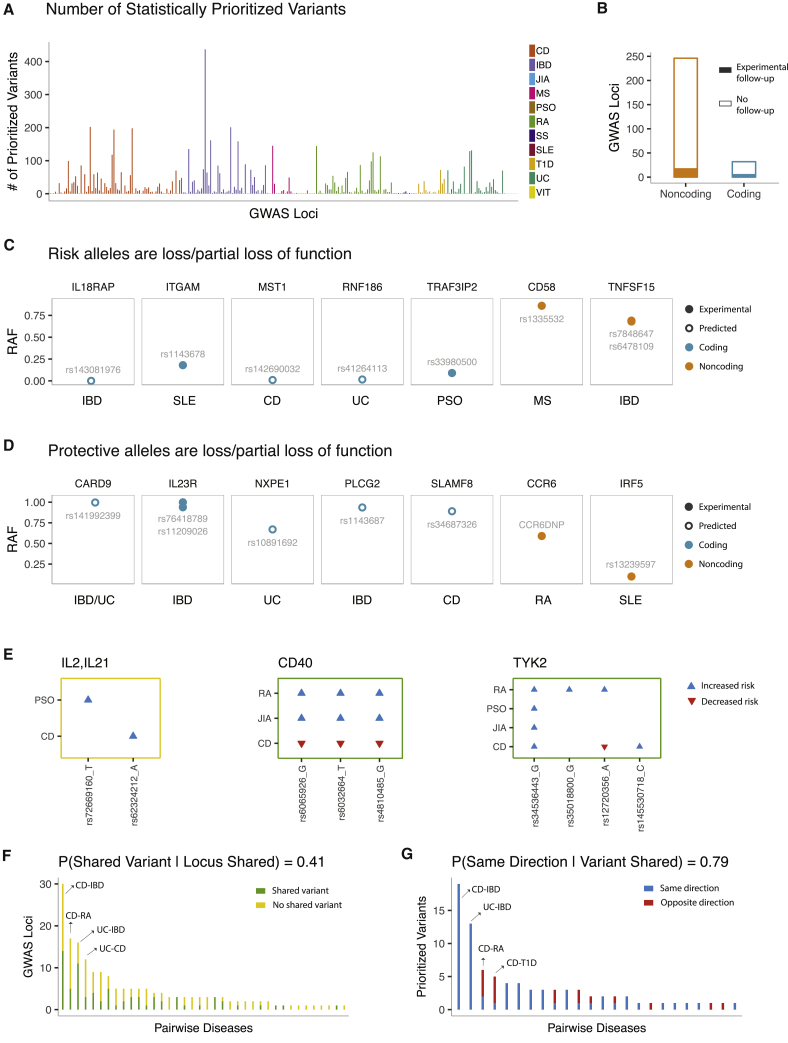
(A) Number of prioritized variants based on statistical estimates across all locus-by-disease combinations.
(B) Locus-by-disease combinations stratified by whether a predicted damaging protein-coding variant was prioritized (n = 32) or not (n = 246). 22 signals (7 protein-coding, 15 non-coding) in total were experimentally followed up on.
(C) Example gene-disease pairs where loss- or partial-loss-of-function alleles increase risk of autoimmune disease.
(D) Example gene-disease pairs where loss- or partial-loss-of-function alleles protect against autoimmune disease.
(E) Example loci with discordance in the set of prioritized variant(s) or direction of genotype effect on the risk of different autoimmune diseases.
(F) The x axis shows the 38 pairwise autoimmune disease combinations for which at least one GWAS locus is shared and fine-mapped for both diseases. The y axis shows the number of loci that harbor at least one shared prioritized variant (green) versus the number of loci that do not harbor any shared prioritized variant (yellow).
(G) The x axis shows the 24 pairwise autoimmune disease combinations that share at least one prioritized variant in at least one locus. The y axis shows the number of shared variant(s) that have the same direction of effect on both diseases (blue) versus the number of variants with opposite direction of effect on each disease (red). Note that when multiple prioritized variants overlapped for two diseases, only one variant per each independent association signal was included during assessment of the direction of effect.