Fig. 3.
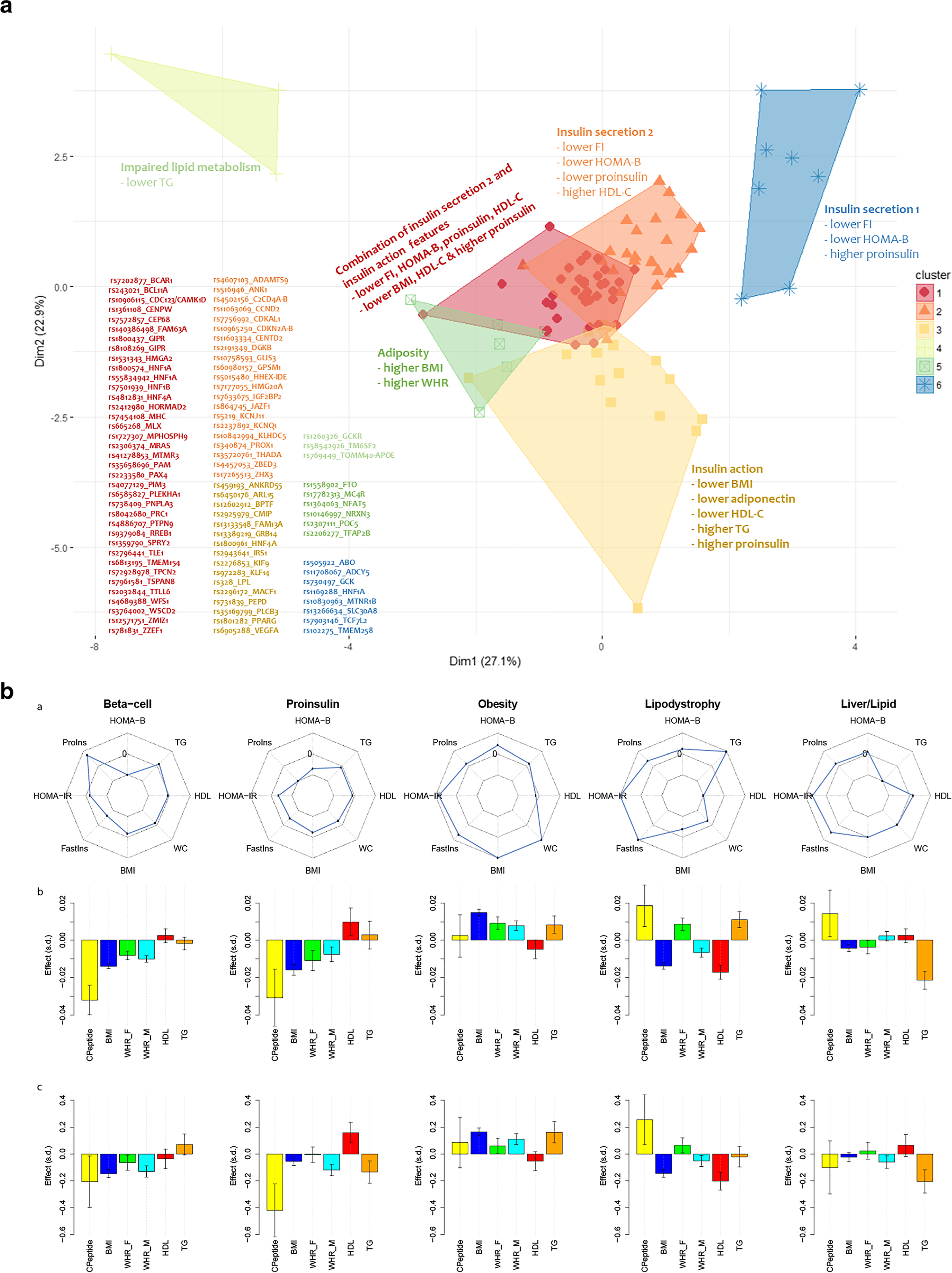
Distinct physiological processes underlying T2D can be defined by genetic variant clusters and “partitioned” polygenic scores. Panel a: clustering variants by direction of effect and magnitude of association with T2D-related metabolic traits defines physiological clusters. Traits like fasting glucose, insulin, lipids or BMI, then further clustering correlated groupings using a fuzzy c-means algorithm that assigns a score to a variant for each cluster (soft clustering, where a variant may be assigned to more than one cluster) produced 5–6 clusters that appeared to represent distinct physiological domains like insulin secretion, insulin action, excess adiposity, or impaired lipid metabolism (panel a is adapted by permission from Springer Nature from Mahajan et al. Nature Genetics. 2018;50(4):559–71) [3]. Panel b: In another study, a similar soft clustering approach defined five distinct physiological domains (columns) with variants combined to generate a “partitioned” polygenic score (PPS). The top row displays spider plots of association of physiological PPSs with metabolic traits (Proins, fasting proinsulin adjusted for fasting insulin; HOMA-B, homeostasis model assessment of beta cell function; TG, serum triglycerides; WC, waist circumference; BMI, body mass index; Fastins, fasting insulin; HOMA-IR, homeostasis model assessment of insulin resistance; WHR-F, waist-hip-ratio in females; WHR-M, waist-hip ratios in males). Points on spider plots in the inner part of the circle show negative association of the PPS and traits and those in the outer circle, positive association. The middle row shows associations of each of five PPSs with metabolic traits in four studies (METSIM, Ashkenazi, Partners Biobank, and UK Biobank, meta-analyzed together), and the bottom row displays the values of individuals with T2D who have each PPS in the highest decile versus all other individuals with T2D. Y-axes are effect size and direction and x-axes are metabolic traits. Soft clustering of genomic data produces genetically defined phenotypes with convincing and distinct patterns, offering potential for genetically based sub-phenotyping of T2D (panel b is adapted from Udler et al. PLoS Medicine. 2018;15(9):e1002654; Creative Commons user license https://creativecommons.org/licenses/by/4.0/)[37]
