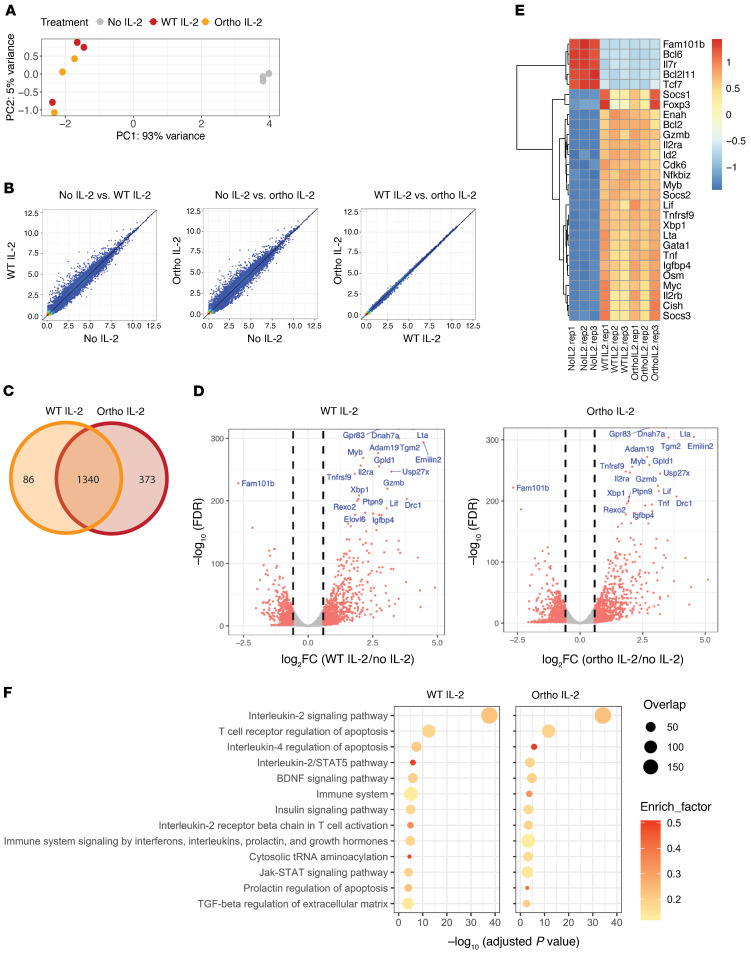
Figure 3. RNA-Seq reveals upregulation of transcripts involved in IL-2 signaling after ortho IL-2 stimulation.
Foxp3GFP+ Tregs were transduced with hEGFR/oIL-2Rβ. hEGFR+ cells were enriched by magnetic cell isolation using an anti-hEGFR mAb. Enriched cells were restimulated with WT or ortho IL-2 for 4 hours after overnight IL-2 starvation. (A) Principal component analysis revealed clustering of WT- (orange) and ortho IL-2–stimulated (red) samples away from unstimulated control cells (no IL-2, gray). (B) Scatterplot of mean RPKM values (log2) in no IL-2 vs. WT IL-2 (left), no IL-2 vs. ortho IL-2 (middle), and WT IL-2 vs. ortho IL-2 (right). (C) Venn diagram showing overlap of DEGs in WT and ortho IL-2–stimulated samples. (D) Volcano plots reveal DEGs in WT- (left) or ortho IL-2–stimulated (right) cells compared with no IL-2. Vertical dashed lines on volcano plots indicate a fold change of ± 1.5. The top 20 most differentially expressed genes are indicated. (E) Heatmap displaying DEGs in IL-2 signaling signature genes among no IL-2–, WT IL-2–, and ortho IL-2–stimulated samples. (F) The top 13 pathways with the smallest adjusted P values in WT IL-2–stimulated cells shown in the BioPlanet 2019 pathway analysis. Pathway analysis of DEGs shown in ortho IL-2–treated cells reveals the same pattern as shown with WT IL-2–treated cells. Plots show the adjusted P values, circle area represents the number of DEGs overlapping with genes in a given pathway, and circle color represents the rich factor reflecting the proportion of DEGs in a given pathway. Data shown are from 1 representative experiment from 3 independent experiments with biological triplicates.