Fig. 2. Up-regulation of N-glycosylation machinery in LDKO livers is dictated by FXR and SHP and not by BA overload.
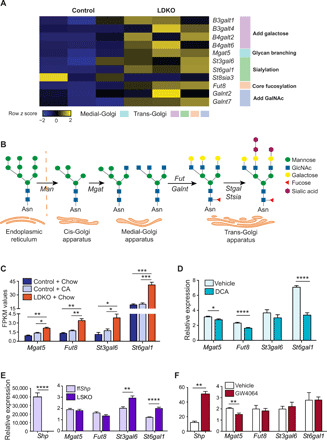
(A) Heatmap indicates up-regulation of key glycosyltransferase genes that are involved in “decorating” glycoproteins in LDKO livers. (n = 3 mice per group, Student t test, fold change > 1.5, P < 0.05). (B) Schematic highlighting the steps of cellular protein processing in ER and Golgi apparatus. (C) Bar graphs comparing the FPKM values indicate up-regulation of glycosylation genes in LDKO livers is not a consequence of BA overload (n = 3 mice per group; one-way ANOVA, *P < 0.05, **P < 0.01, and ***P < 0.001). (D) Incubation of primary hepatocytes with a toxic BA, such as DCA, suppresses the N-glycosylation genes instead (n = hepatocytes cultured in triplicates from two mice per group; Student t test, *P < 0.05 and ****P < 0.0001). Qualitative RT-PCR analysis of (E) liver-specific SHP knockout (LSKO) livers and (F) FXR-activated (treated with GW4064, FXR agonist) livers reveal that FXR and SHP individually regulate some of these glycosylation genes (n = 5 to 8 mice per group; Student t test, **P < 0.01 and ****P < 0.0001).
