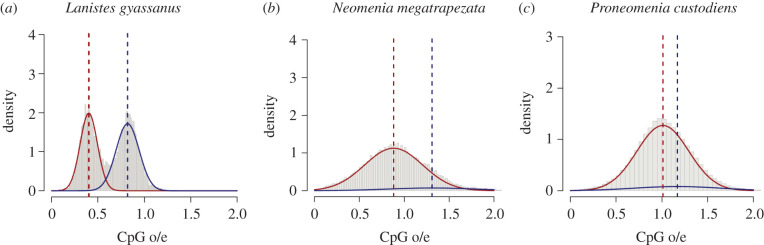
Figure 2.
Examples of CpG o/e distribution patterns in protein-coding sequences in three mollusc species. The red and blue curves represent the two Gaussian distributions fitted to the data. The correspondingly coloured dashed lines represent the mean values of the fitted distributions. (a) Lanistes nyassanus shows a typical bimodal CpG o/e distribution with a portion that has low CpG o/e values (sequences mainly affected by CpG depletion) and one that has high CpG o/e values (sequences less affected by CpG depletion) (b) Neomenia megatrapezata displays a ‘unimodal, indicative of methylation' CpG o/e distribution. This distribution lacks clear bimodality, but their low CpG o/e component has a characteristically large tail containing a significant amount of data. (c) Proneomenia custodiens shows a ‘unimodal, not indicative of DNA methylation' CpG o/e distribution. The mean values of the two fitted distributions are almost identical and the proportion of data belonging to the smaller component is small. (Online version in colour.)