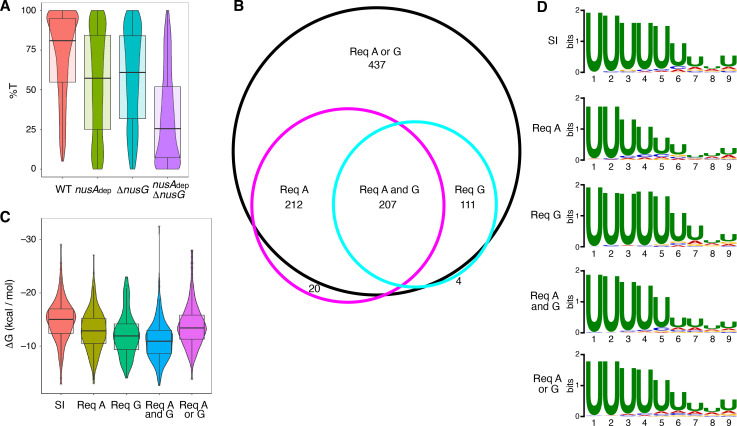
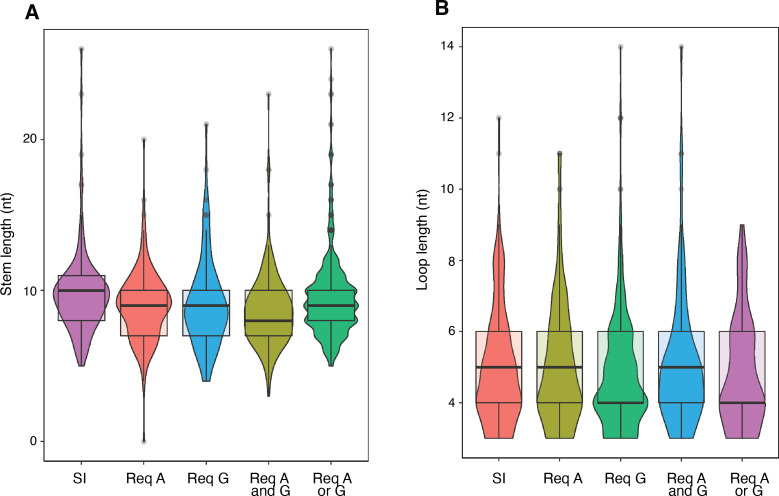
Figure 1. NusG is an intrinsic termination factor that works with NusA to stimulate suboptimal terminators.
(A) Violin plots overlayed with box plots showing the distribution of termination efficiency (%T) in wild-type (WT), nusAdep, ΔnusG, and nusAdep ΔnusG strains. For this and all box plots, boundaries of the box designate the interquartile range (IQR), while upper/lower whiskers extend from the 75th/25th percentile to the largest/smallest value no further than 1.5*IQR in either direction. All pairwise p-values can be obtained from Supplementary file 3. (B) Venn diagram showing the number and overlap of terminators that were classified as being dependent (Δ%T ≥ 25) on NusA (Req A) and/or NusG (Req G). The black circle contains all terminators that were classified as dependent in the nusAdep ΔnusG strain, the magenta circle contains all terminators that were classified as dependent in the nusAdep strain, and the cyan circle contains all terminators that were classified as dependent in the ΔnusG strain. Intrinsic terminator subpopulations that require NusA and/or NusG in any fashion to terminate efficiently are specified. (C) Violin plots overlayed with box plots showing the distribution of predicted hairpin strength as reported in ΔG (kcal/mol) for all identified subpopulations including the strong and independent (SI) terminators. All pairwise p-values can be obtained from Supplementary file 3. (D) Sequence logos of the U-rich tracts generated from the nine nucleotide (nt) window downstream of the predicted hairpins for all identified subpopulations including the SI terminators. All pairwise comparisons can be found in Supplementary file 3.