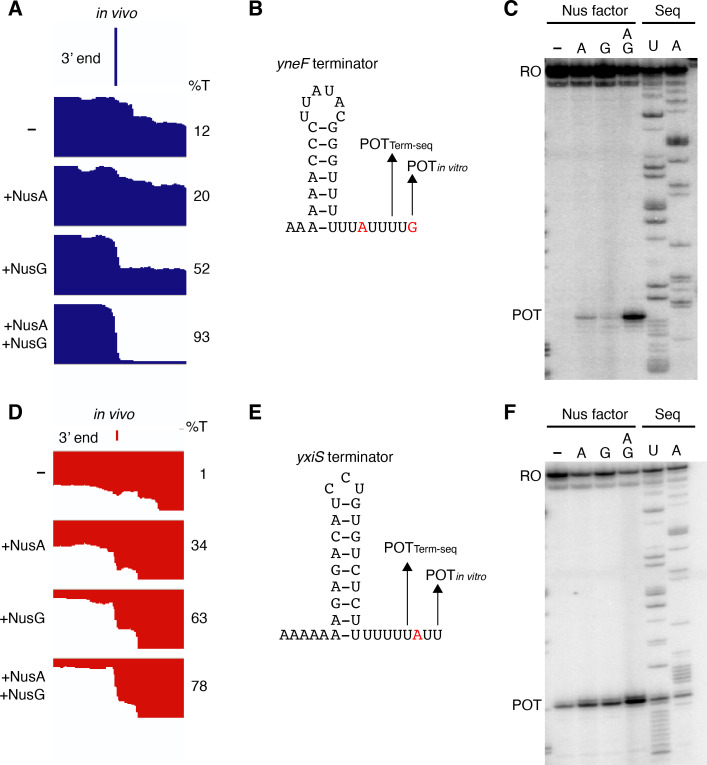
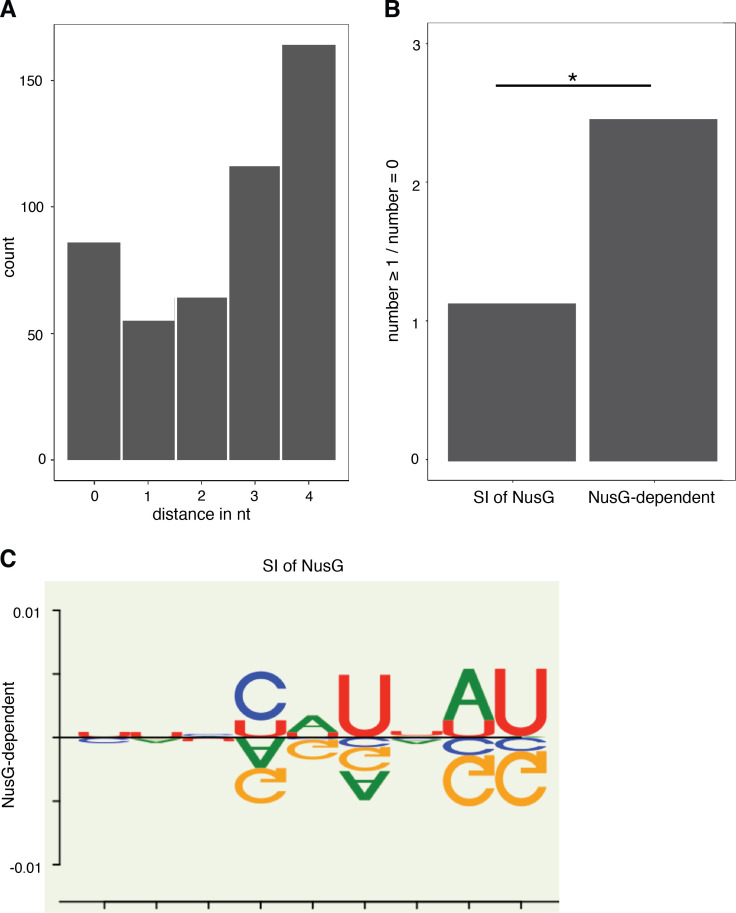
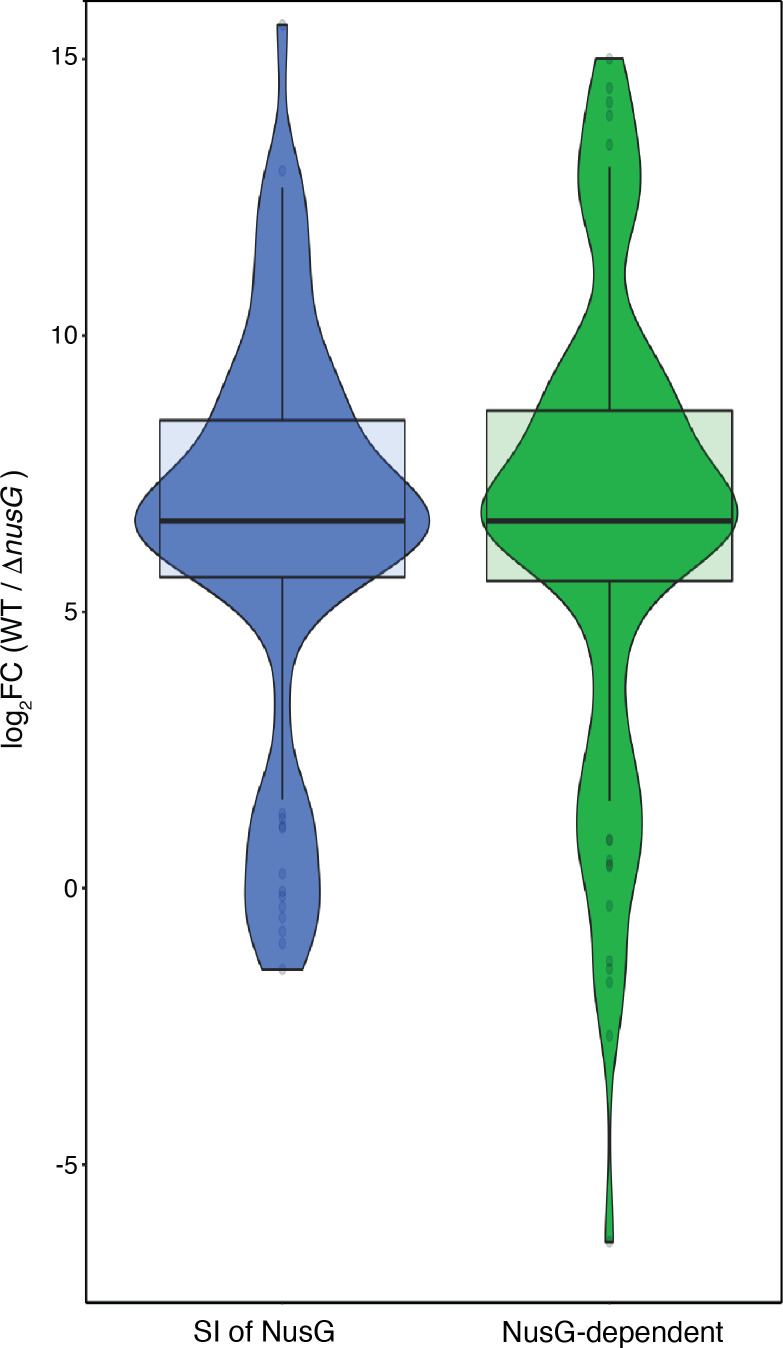
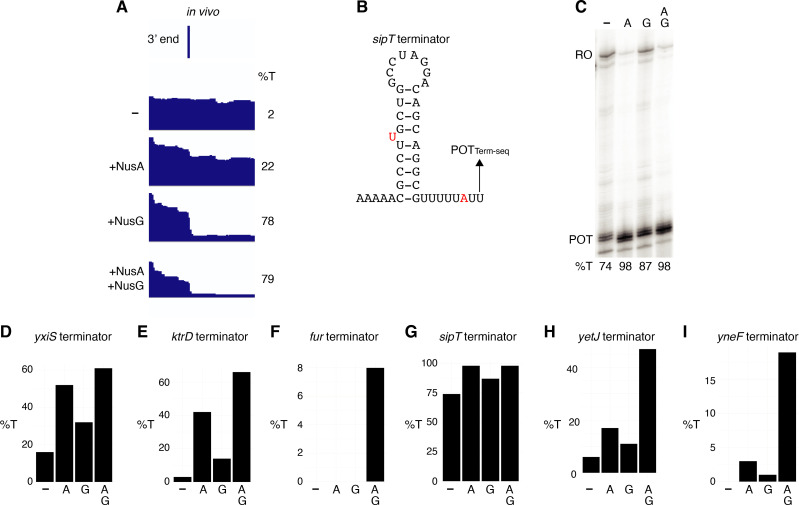
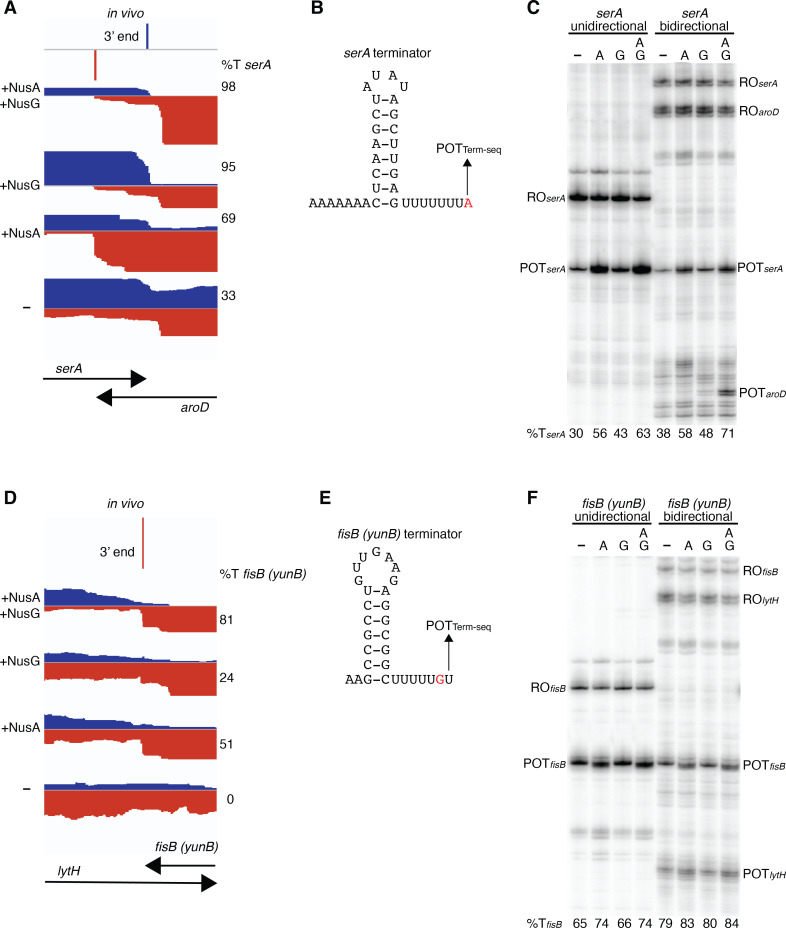
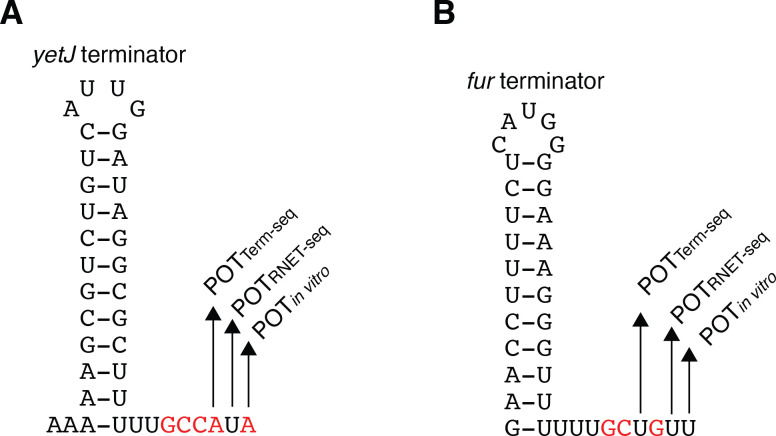
Figure 4. NusG stimulates terminators with particularly weak terminal base pairings.
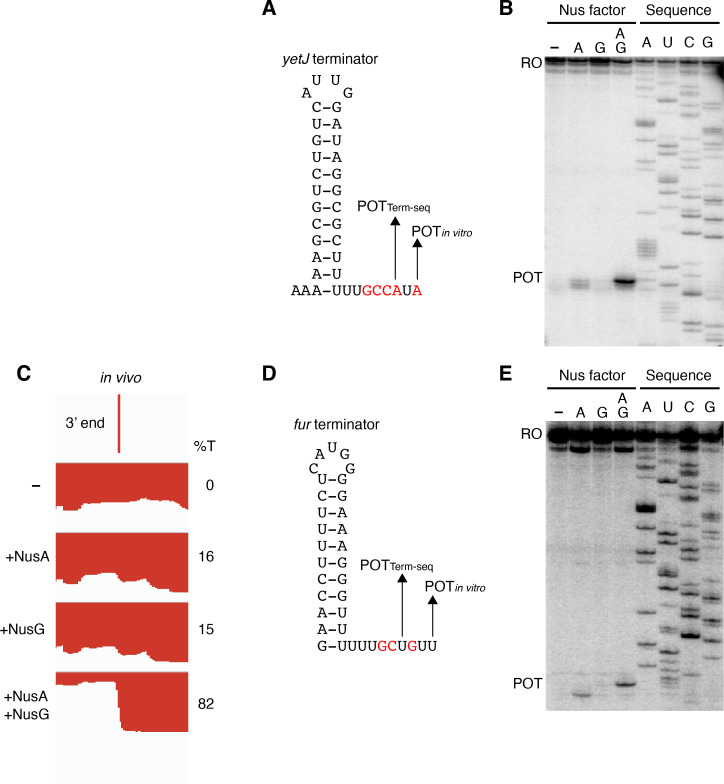
(A) YetJ terminator showing the point of termination identified in vivo by Term-seq (POTTerm-seq) and by in vitro transcription in the +A+G condition (POTin vitro). Disruptions in the U-rich tract are shown in red. The upstream A tract is also shown. An IGV screenshot of this terminator is shown in Figure 2A. (B) Single-round in vitro termination assay with the yetJ terminator. Experiments were performed in the absence (–) or presence of NusA (A) and/or NusG (G) as indicated. Positions of terminated (POT) and run-off (RO) transcripts are marked. RNA sequencing lanes (A, U, C, G) are labeled. (C) IGV screenshot of a genomic window centered around the fur terminator. Top track is the 3’ end identified by Term-seq. Bottom tracks are the RNA-seq coverage data for the nusAdep ΔnusG (-), ΔnusG (+NusA), nusAdep (+NusG), and WT (+NusA +NusG) strains. %T in each strain is shown on the right of each track. Transcription proceeds from right to left. (D, E) Identical to panels (A, B) except that it is the fur terminator. (D) Note that the terminal three base pairs contain the A tract and one G residue.