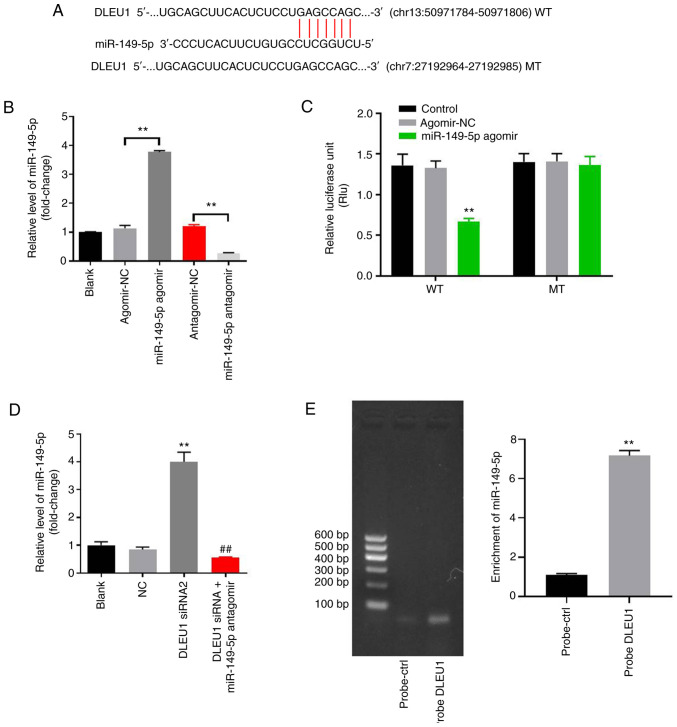
Figure 3.
DLEU1 can sponge miR-149-5p in OSCC. (A) Gene structure of DLEU1 indicated the binding site of miR-149-5p in its 3′UTR. (B) OSCC cells were transfected with antagomir/agomir NC, miR-149-5p agomir/antagomir for 24 h. Then, the expression of miR-149-5p in OSCC cells was detected via RT-qPCR. (C) The relative luciferase activity in Cal-27 cells after co-transfection with miR-149-5p and WT/MT DLEU1 3′UTR was measured using a dual-luciferase reporter assay. (D) Cal-27 cells were treated with NC, DLEU1 siRNA2 or DLEU1 siRNA2 + miR-149-5p. Then, the expression of miR-149-5p in Cal-27 cells was detected via RT-qPCR. (E) The association between DLEU1 and miR-149-5p was explored using an RNA pull-down assay. **P<0.01 vs. control; ##P<0.01 vs. DLEU1 siRNA2. DLEU1, deleted in lymphocytic leukemia 1; OSCC, oral squamous cell carcinoma; NC, negative control; siRNA, small interfering RNA; miR, microRNA; UTR, untranslated region; WT, wild-type; MT, mutant; RT-qPCR, reverse transcription-quantitative PCR.