Figure 1.
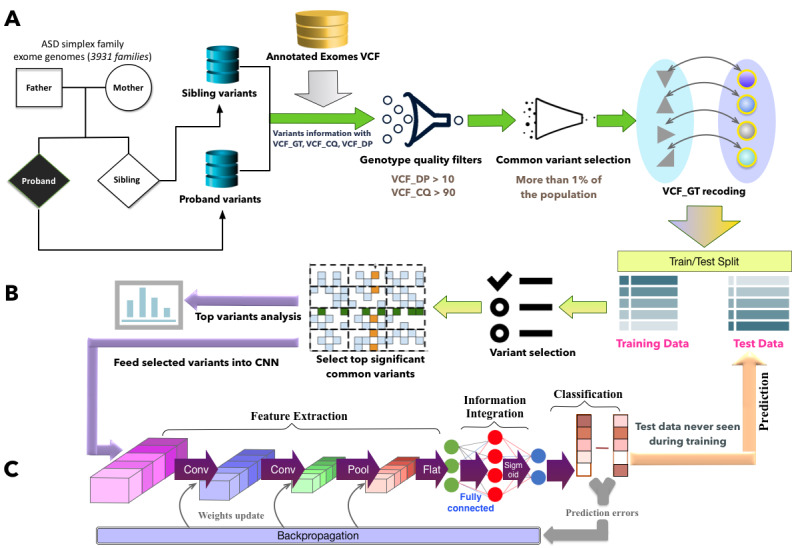
Overall framework for deciphering contributory common variants and predicting autism spectrum disorder diagnosis. A. Data preprocessing. VCF_GT recoding is to encode VCF_GT values as dummy variables. If both alleles are reference alleles, it is encoded as 0; if both alleles are alternate alleles, it is encoded as 2; otherwise, it is 1. B. Data split and significant variant selection. The data set was split into training set and test set. Variants were ranked based on their chi-score and P value, and only top ranked (high chi-score value and low P value) variants were selected as contributory common variants for autism spectrum disorder. C. Convolutional neural network classifier. The selected significant common variants in the training data were fed into a convolutional neural network to train a classifier. Thereafter, the trained model was applied on the test data for autism spectrum disorder diagnosis prediction. ASD: autism spectrum disorder; CNN: convolutional neural network; SSC: Simons Simplex Collection; VCF: variant call format; VCF_CQ: variant call format-conditional genotype quality; VCF_DP: variant call format-read depth; VCF_GT: variant call format-genotype quality.
