Figure 3.
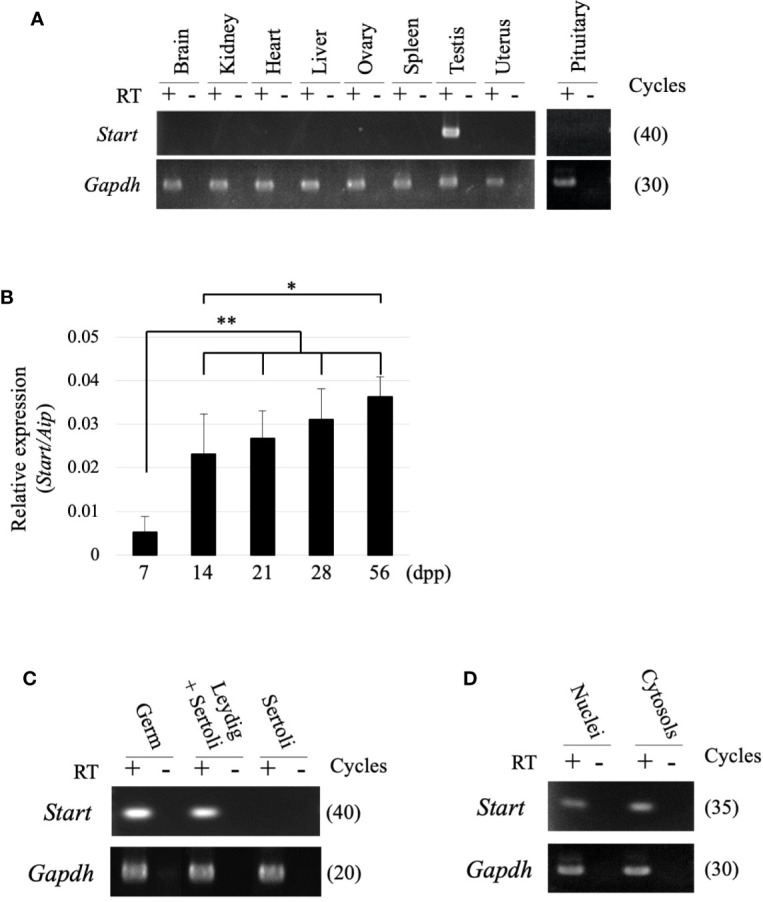
Expression of the Start transcript. (A) Tissue specificity of Start. Total RNAs from nine mouse tissues, as indicated, were collected from adult wild-type mice and used for RT-PCR with a random hexamer for reverse transcription. The Gapdh gene was detected as an internal control. The cycle number of each PCR is shown in parenthesis. We obtained similar results with the three biological replicates, and a representative result of the three experiments is shown. (B) Start expression during testis development. Mouse testes at 7, 14, 21, 28, and 56 dpp were collected, and total RNAs were purified and used for qRT-PCR. The Aip gene was used as an internal control to normalize the Start expression level. Data are presented as means ± S.D. from three independent experiments with three biological replicates. Statistical significance was analyzed by Tukey’s HSD test. *P < 0.05, **P < 0.01. (C) RT-PCR with fractionated testicular cells. Total RNAs from germ cells, Leydig/Sertoli cells, and Sertoli cells were collected and used for RT-PCR. The Gapdh gene was utilized as an internal control. The cycle number of each PCR is shown in parenthesis. (D) RT-PCR for subcellular localization of Start. Total RNAs were purified from nuclear and cytosolic fractions of germ cells and used for RT-PCR. Gapdh was used to amplify a region within exon 6 as an internal control. The cycle number is indicated in parenthesis.
