Fig. 1.
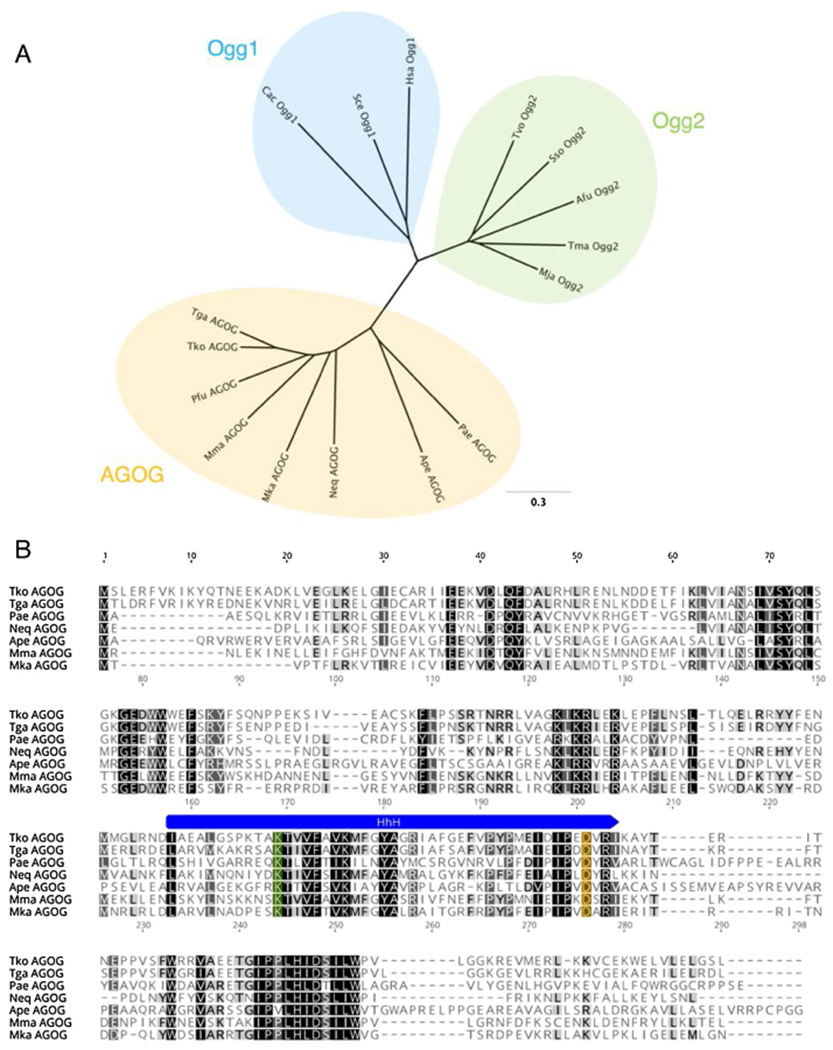
T. kodakarensis AGOG is a member of the AGOG subfamily of Ogg enzymes. A, Unrooted phylogenetic tree of Ogg family enzymes with the 3 distinct subfamilies highlighted: Ogg1 (blue), Ogg2 (green), and AGOG (yellow). The tree was generated using Geneious v11.0.2. B, Amino acid sequence alignment of members of the AGOG subfamily. The conserved Helix-hairpin-Helix (HhH) is marked in blue, the catalytic lysine residue in orange, and the active site aspartic acid in green. The species abbreviations and GenBank ascension numbers used are as follows: Archaea: Afu, Archaeoglobus fulgidus (Ogg2, AAB90876); Ape, Aeropyrum pernix (AGOG, BAA79686); Mja, Methanocaldococcus jannaschii (Ogg2, AAB98720); Mma, Methanococcus maripaludis (AGOG, CAF29860); Neq, Nanoarchaeum equitans (AGOG, AAR39356); Mka, Methanopyrus kandleri (AGOG, AAM01756); Pae, Pyrobaculum aerophilum (AGOG, AAL64050); Pfu, Pyrococcus furiosus (AGOG, AAL81028); Sso, Saccharolobus solfataricus (Ogg2, AAK41186); Tga. Thermococcus gammatolerans (AGOG, ACS34155); Tko, Thermococcus kodakarensis (AGOG, BAD85129); Tvo, Thermoplasm volcanium (Ogg2, WP_010916318); Bacteria: Cac, Clostridium acetobutylicym (Ogg1, NP_349313); Tma, Thermotoga maritima (Ogg2, AGL50755); Eukarya: Hsa, Homo sapiens (Ogg1, NP_002533); Sce, Saccharomyces cerevisiae (Ogg1, NP_013651).
