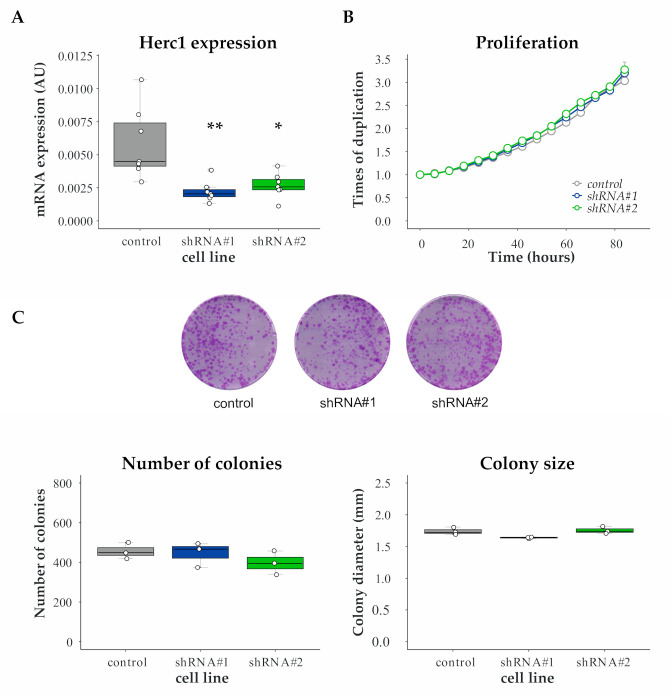
Figure 2.
Proliferation is unaffected by HERC1 depletion. MDA-MB-231 cells were stably transduced with an empty vector (control) or two different shRNAs (#1 and #2) targeting HERC1. (A) Efficiencies of shRNA-mediated target gene knockdown were confirmed by RT-PCR (top, n = 7, one-way ANOVA, Dunnett’s multiple comparison test; shRNA #1 p = 0.0014 (**) and shRNA #2 p = 0.0365 (*)). (B) An area-based microscopy method was used to determine cell growth over time. Cells were seeded onto wells and allowed to attach. At the indicated time points, cells were photographed, and the occupied area was calculated. The graph shows the occupied area relative to time = 0 h. Doubling times = control, 47.01 ± 0.2087; shRNA #1, 46.23 ± 0.3667; shRNA #2, 47.38 ± 1.023 (n = 3, Kruskal–Wallis, Dunn’s multiple comparison test; shRNA #1 p > 0.9999 and shRNA #2 p = 0.4503). (C) Cells were subjected to colony formation assay: (Top) representative images of the crystal violet-stained 100 mm dishes at the end of the experiment; (Bottom Left) number of colonies generated after 15 days (n = 3, Kruskal–Wallis, Dunn’s multiple comparison test; shRNA #1 p > 0.9999 and shRNA #2 p = 0.4661); and (Bottom Right) diameter of the resulting colonies (n = 3, Kruskal–Wallis, Dunn’s multiple comparison test; shRNA #1 p = 0.1473 and shRNA #2 p > 0.9999).