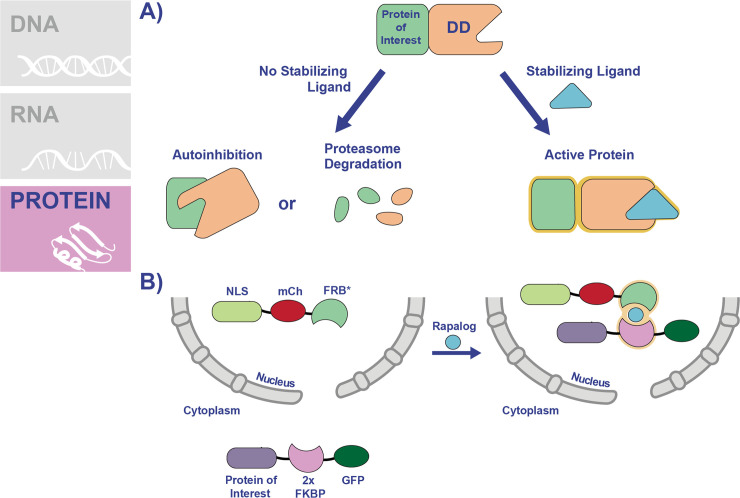
Fig 4. Conditional knockdown of protein function using degradation or KS systems.
(A) The FKBP-DD and DD systems work using a similar mechanism, thus a combined schematic of how these systems work is shown. For simplicity, both the FKBP-DD and DD are depicted and referred to as DD. The sequence encoding the DD is inserted into the genome in frame immediately following the gene of interest. The DD contains point mutations which render the protein domain unstable, causing the protein to be recognized for degradation by the proteasome. In the case of chaperone proteins appended to the DD, the unfolded domain is recognized by the chaperone, which results in autoinhibition. The DD is stabilized through the addition of a ligand (TMP for DD and Shld1 for FKBP-DD). Addition of the ligand stabilizes the DD to prevent its degradation by the proteasome or autoinhibition. (B) The KS system allows for the conditional mislocalization of proteins to inhibit their functions. In this system, the protein of interest is fused to 2 copies of the FKBP domain in a parasite line that expresses an FRB* “mislocalizer” fusion protein, engineered to localize to the nucleus or plasma membrane. Addition of rapamycin (or rapalog) dimerizes the FKBP/FRB* domains, resulting in the relocalization of the protein of interest to one of these sites. DD, destabilization domain; FKBP, FK506 binding protein 12; FKBP-DD, FK506 binding protein destabilization domain; FRB, FKBP12-rapamycin-binding; GFP, green fluorescent protein; KS, knock sideways; mCh, mCherry; NLS, nuclear localization signal; TMP, trimethoprim.