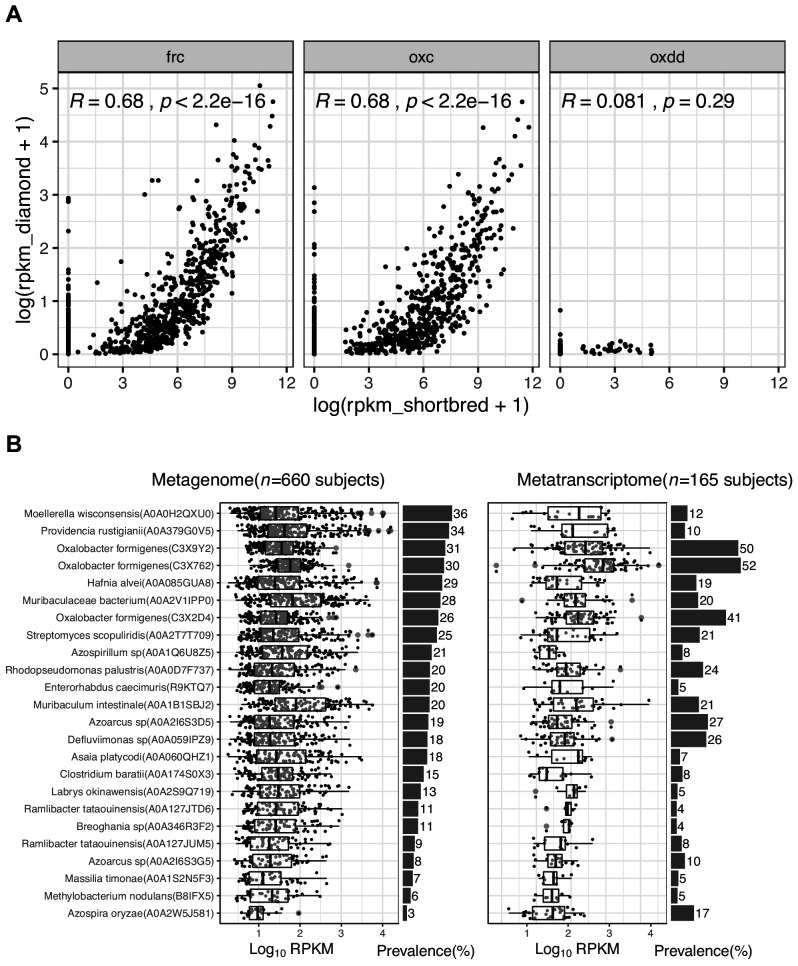
Figure 3. Expression of type II ODP of microbial species within the intestinal microbiota of healthy humans.
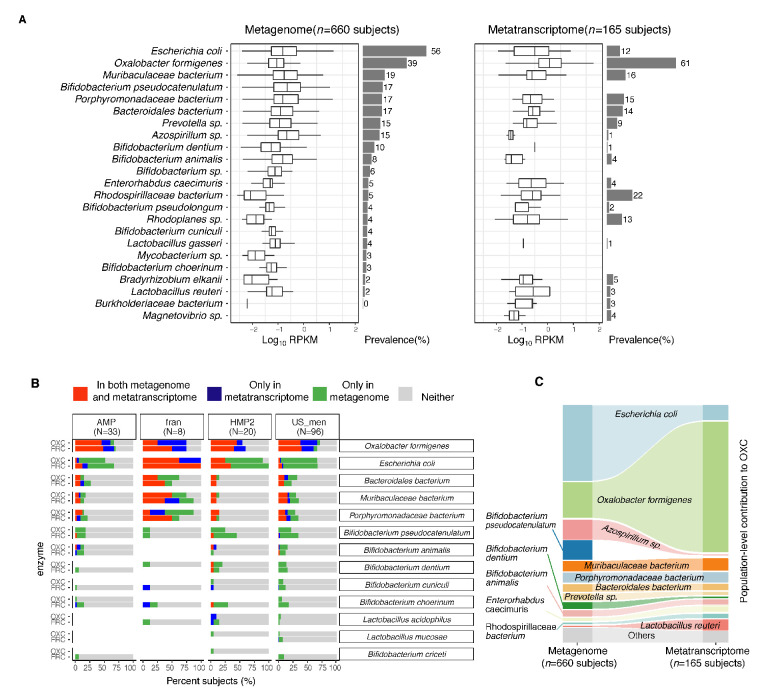
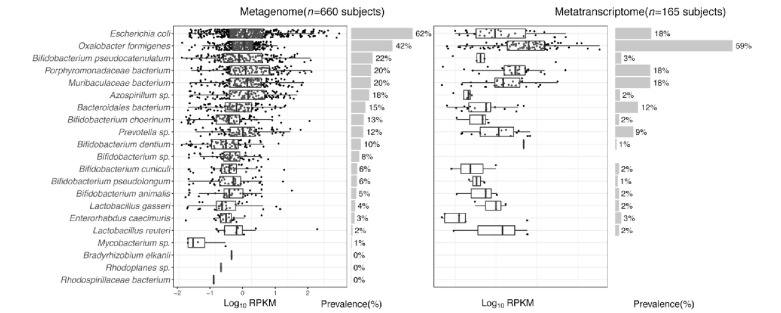
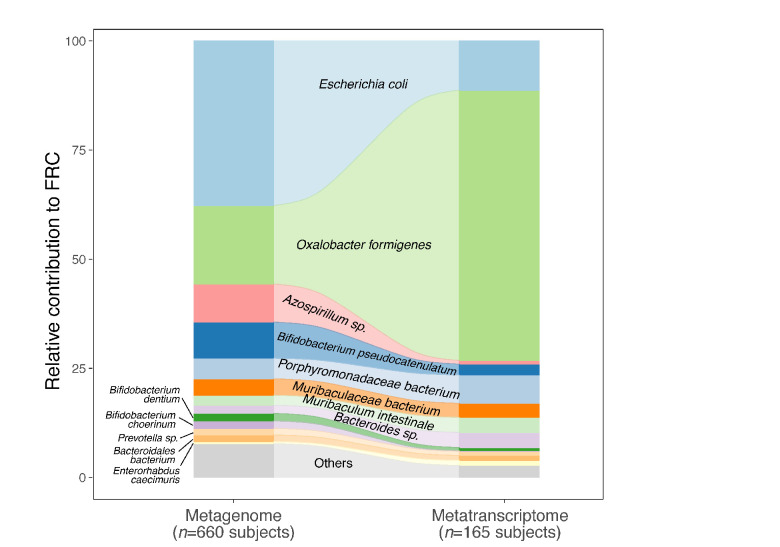
(A) Abundance and prevalence of OXC of microbial species in the metagenome (left) or metatranscriptome (right) of 660 and 165 subjects. Box plots indicate the abundance of microbial OXC (log10 RPKM) among subjects in whom OXC is detected, and are generated with ggplot2 with outliers excluded. Bar plots indicate the prevalence of microbial oxc, with percentage annotated. Microbial species are ordered by the corresponding metagenomic OXC prevalence. A parallel analysis for FRC is shown in Figure 3—figure supplement 5. (B) Detection of OXC and FRC of microbial species in the subject-matched metagenome and metatranscriptome, by study. For each microbial ODE, the subjects are divided into four groups (shown in different colors) based on the co-detection of ODE in the matched metagenome and metatranscriptome, with percent (%) of which reflected. The fran Study, from which E. coli ODP was detected in all subjects, used a sample extraction method known to induce E. coli, as noted in their publication (Franzosa et al., 2014). (C) Population-level contribution of individual species to metagenomic (left) or metatranscriptomic (right) OXC. The population-level contribution of each species was calculated at a relative scale (see Materials and methods) and plotted. Raw values can be found in Supplementary file 1a. The 10 species that have the highest metagenomic or metatranscriptomic contribution are shown. A parallel analysis for FRC is shown in Figure 3—figure supplement 3.