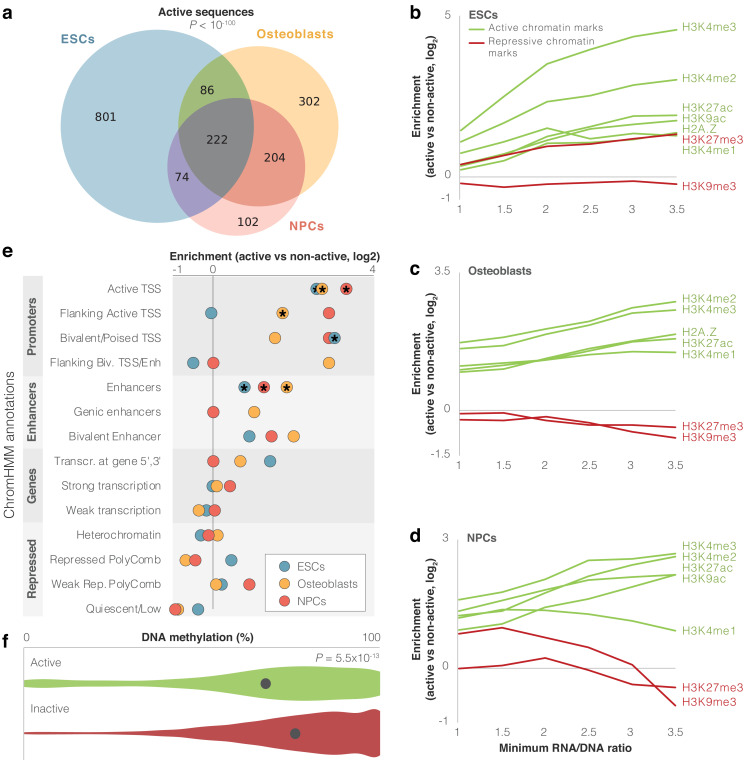
Figure 2. Identification of modern human sequences promoting expression in lentivirus-based MPRA (lentiMPRA).
(a) Overlap between cell types of active sequences. Super Exact test p-value is shown for the overlap of the three groups. (b-d) Enrichment levels of active and repressive histone modification marks within active sequences. Enrichment is computed compared to inactive sequences. The enrichment of H3K27me3 in embryonic stem cells (ESCs) possibly reflects the presence of this mark in bivalent genes, which become active in later stages of development (Blanco et al., 2020). For confidence intervals, see Supplementary file 2. (e) Enrichment of differentially active sequences in various chromatin-based genomic annotations. Missing circles reflect no differentially active sequences in that category. Stars mark significant enrichments (false discovery rate [FDR] <0.05). (f) Violin plots of DNA methylation levels for active (green) vs. inactive (red) sequences in osteoblasts. Methylation levels per sequence were computed as the mean methylation across all modern and archaic human bone methylation samples. The circle marks mean methylation across all sequences in each group. t-test p-value is shown.