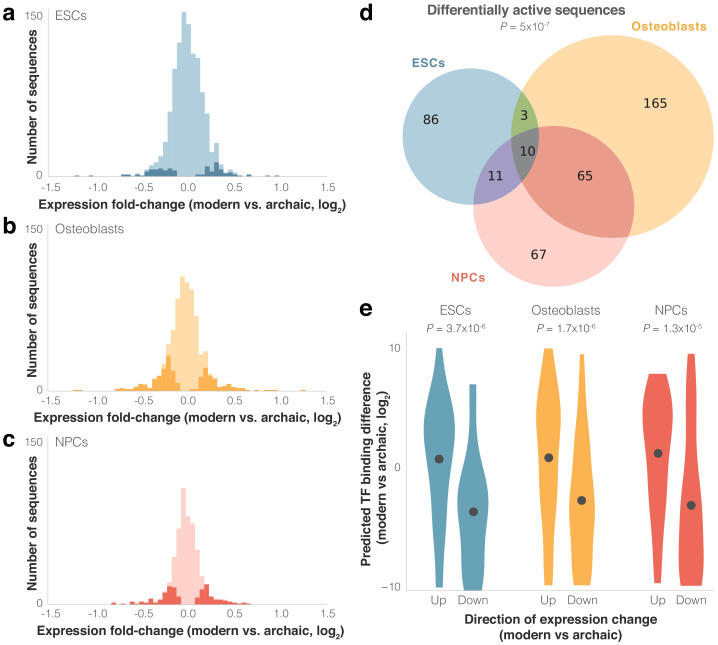
Figure 3. Differential activity of derived modern human sequences.
(a–c) Distributions of expression fold-changes (RNA/DNA) of active (light) and differentially active (dark) sequences in each cell type. (d) Overlap of differentially active sequences between cell types. Super Exact test p-value is presented for the overlap of the three groups compared to active sequences. In the 10 sequences that were differentially active across all three cells types, the direction of fold-change was identical across all cell types (p=1.9×10−3, binomial test). (e) Violin plots of predicted transcription factor (TF) binding score difference between modern and archaic sequences. Positive scores represent increased binding in the modern sequence. Points show mean.