Figure 3.
Alphacoronavirus HCoV-229E produces classes of TRS-independent sgRNA that are abundantly expressed
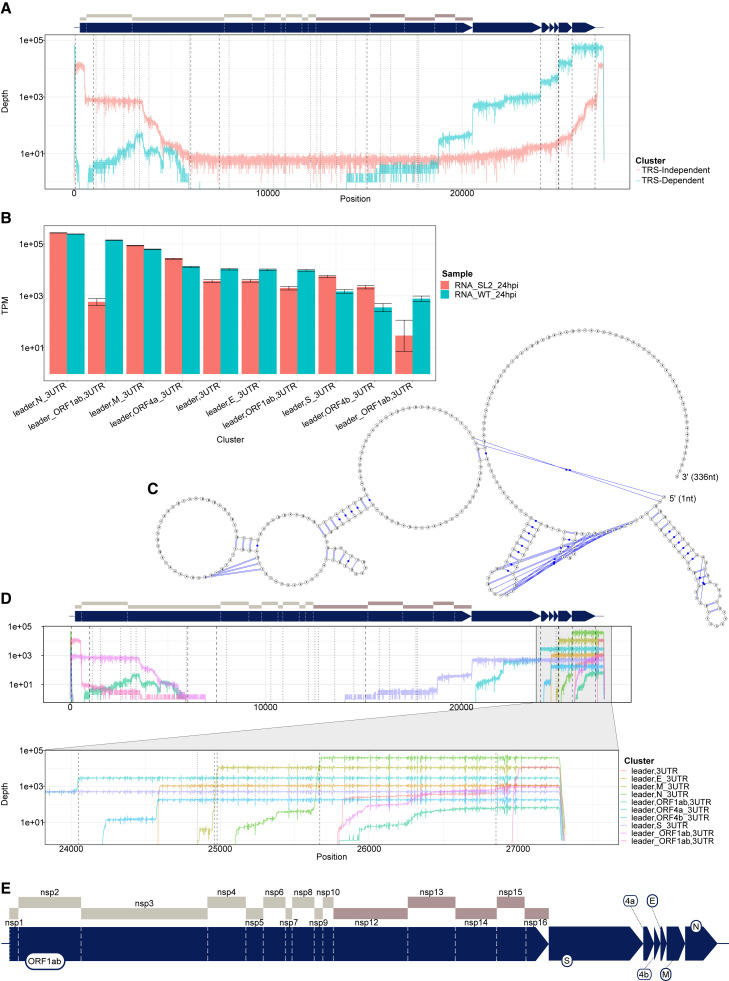
The transcript nomenclature W_X,Y_Z indicates that the transcript consists of the segment from W to X joined with the segment from Y to Z.
(A) Total depth of TRS-independent versus TRS-dependent sgRNA in HCoV-229E. Dashed vertical lines indicate positions of TRS motifs, with alternative motifs detected using FIMO shown in dotted and dot-dash lines.
(B) Normalized transcript counts (TPM viral-mapped reads) of major sgRNA from HCoV-229E for wild-type (WT) or with stem loop 2 replaced (SL2). Error bars indicate 95% binomial CI of TPM estimate using the logistic parameterization.
(C) Predicted secondary structure of ORF10 + 3′ UTR from SARS-CoV-2 showing bulged stem loop in ORF10. Prediction calculated with IPknot software.
(D) Transcript coverage of major classes of sgRNA in terms of total read depth across all cDNA samples, shown on log scale. Dotted lines indicate positions of TRS.
(E) Enlarged schematic of genome annotation for HCoV-229E. Regions are to scale.
See also Figure S1.