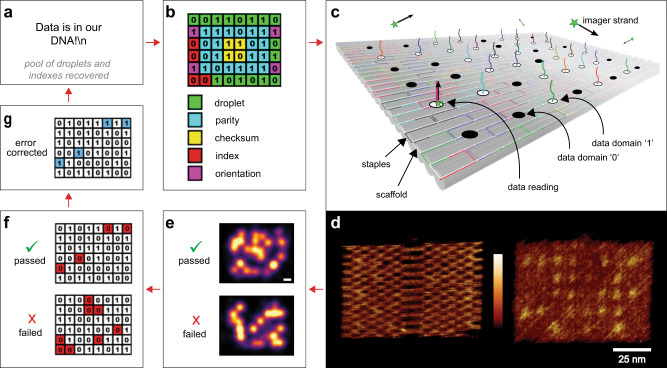
Fig. 1. Binary dNAM overview.
The test message (a) for optically reading dNAM was ‘Data is in our DNA!’. The message was encoded and then synthesized into 15 dNAM origami. For clarity, only one of the 15 designs is shown in (b). The data domain colors correspond to their bit values as follows: droplet (green), parity (blue), checksum (yellow), index (red), and orientation (magenta). Site-specific localization is enabled by extending or not-extending the structural staple strands of the origami to create physical representations of 1s and 0s. The presence, absence, and identity of a data strand’s docking sequence defines the state of each data strand and is assessed by monitoring the binding of data imager strands via DNA-PAINT in (c). AFM images of an origami nanostructure are depicted in (d), with both the expected raft honeycomb structure (left) and data strands (right) visible. The scale bar is 25 nmin the AFM images and the color scale ranges from 0–1 nm in height. To ‘read’ the encoded message, 4 μL of the DNA origami mixture, containing 0.33 nM of each origami, was imaged via DNA-PAINT. Two representative origami cropped from the final rendered image are shown in (e), scale bar, 10 nm. All structures identified as origami in the rendered image were converted to a matrix of 1’s and 0’s corresponding to the pattern of localizations seen at each data domain in (f). The red boxes in (f) now indicate errors. The decoding algorithm performed error correction where possible in (g) and successfully retrieved the entire message when sufficient data droplets and indexes were recovered in (a). The blue boxes in (g) now indicate corrected errors.