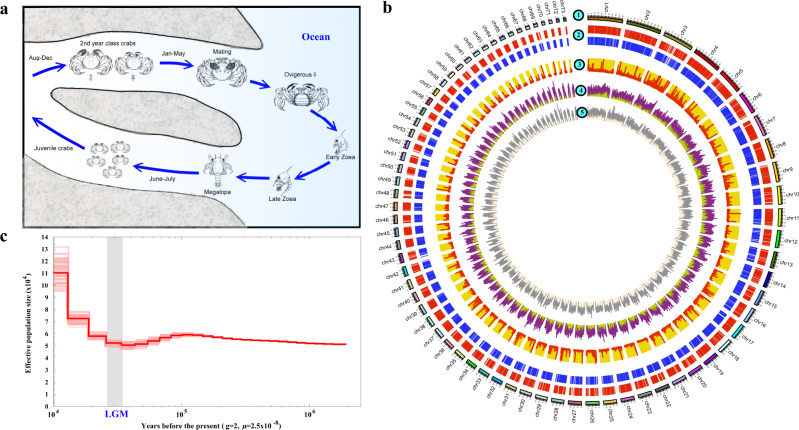
Fig. 1. Characterization of the Eriocheir sinensis genome.
a The life cycle of E. sinensis. Adult crabs attain sexual maturity in estuaries followed by egg hatching and then upstream migration of megalopae to rivers and lakes. b The genome landscape of E. sinensis. From outer to inner circles: b1, marker distribution on 73 linkage groups at Mb scale; b2, protein-coding genes located on anchored scaffolds; b3, distribution of high- (>60%) and low-GC (<20%) contents in 200-bp regions across the genome within a 100-kb sliding window; b4, microsatellite distribution (1–6 bp) across the genome drawn in 100-kb sliding windows; b5, SNP density of resequenced individuals drawn in 100-kb sliding windows within a 50-kb step, with polymorphism hotspot regions (P < 0.05) colored in red. The genomic visualization was created using the program Circos. c Demographic histories of mitten crab reconstructed using PSMC model. The period of the last glacial maximum (LGM, ~18,000–22,000 years ago) is shown as a gray bar. g (generation time) = 2 years; µ (neutral mutation rate per generation) = 2.5 × 10−8.