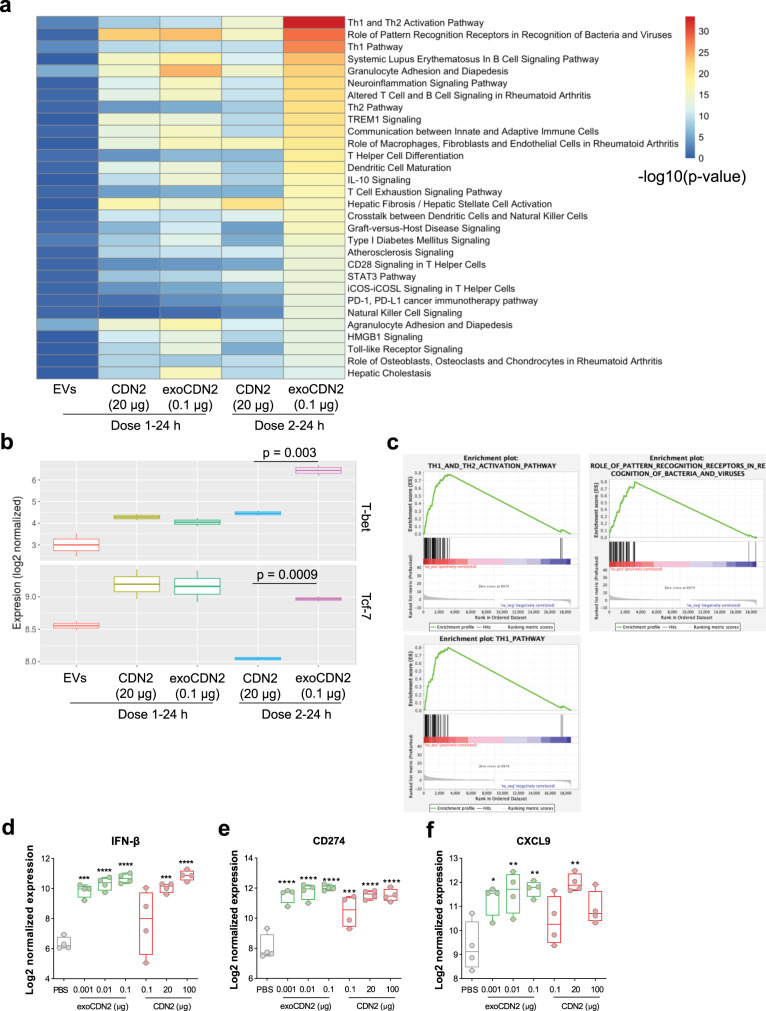
Fig. 5. exoSTING induced interferon stimulated gene signatures.
a Comparative pathway analysis of the global gene expression changes analyzed by RNA sequencing, 24 h after 1 or 2 IT injections of PBS, unloaded EVs, CDN2 (20 µg), and exoCDN2 (0.1 µg) into B16F10 tumors (n = 2 biological replicates per group). The data is representative of five of biological replicate samples in each treatment group. All treatments were compared to PBS treatment for gene expression. b Normalized expression level of Th1 transcription factors, T-bet and Tcf7. Adjusted P values are indicated. c Gene Set Enrichment Analysis of three gene sets that are upregulated by exoCDN2. d–f Four hours after IT injection of PBS, CDN2 (0.1, 20, or 100 µg), and exoCDN2 (0.001, 0.01, or 0.1 µg) into B16F10 tumors, RNAs were purified from tumors (n = 4 animals per group) and differentially expressed genes were analyzed using NanoString technology. Relative expression of IFN-β (d), CD274 (e), and CXCL9 (f) measured by NanoString. Data are presented as means ± s.e.m from replicate samples as indicated. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by one-way ANOVA with Tukey’s multiple comparison test.