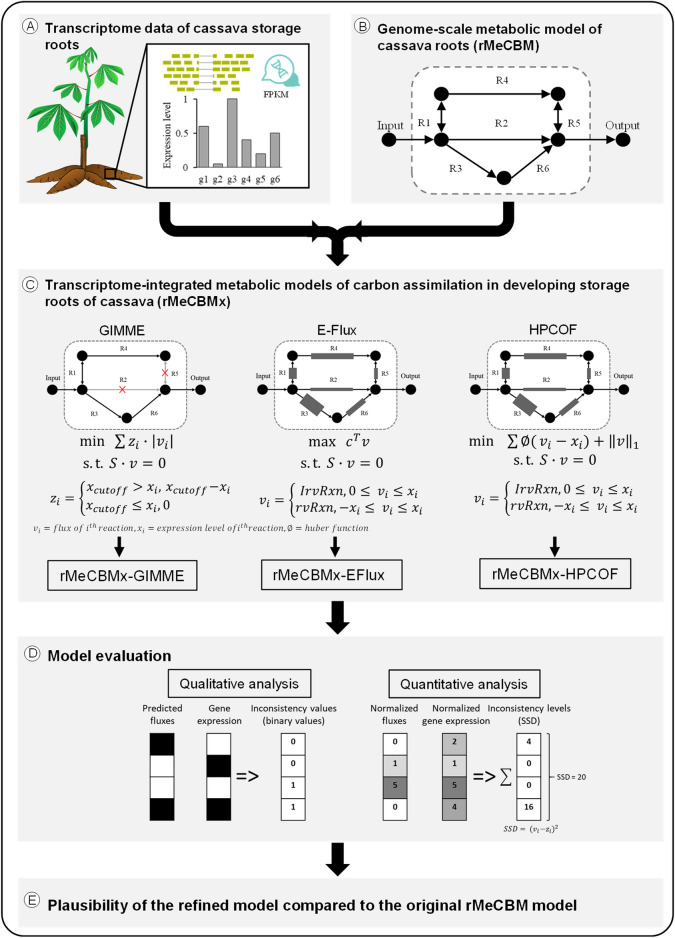
Figure 1.
Overall methodology. (A) Transcriptome data analysis of gene expression in developing storage roots of cassava. (B) Genome-scale metabolic model of cassava storage roots, rMeCBM, obtained from Chiewchankaset et al.16. (C) Development of transcriptome-constrained GSMM models through GIMME, E-Flux, and HPCOF methods. Metabolic fluxes of each reaction were constrained by the relative expression level of the highest expressed genes responsible for the reaction. (D) Evaluation of model performance based on the consistency of predicted fluxes with gene expression levels, both qualitatively and quantitatively. (E) Plausibility testing of the transcriptome-constrained GSMM model by simulation of storage root growth of the high-yielding cultivar CMC-9, sensitivity analysis, and analysis of the consistency of predicted flux distribution with the gene expression data.