Figure 1.
An optimized workflow for full-length cDNA library construction from eukaryotic tRNA pools
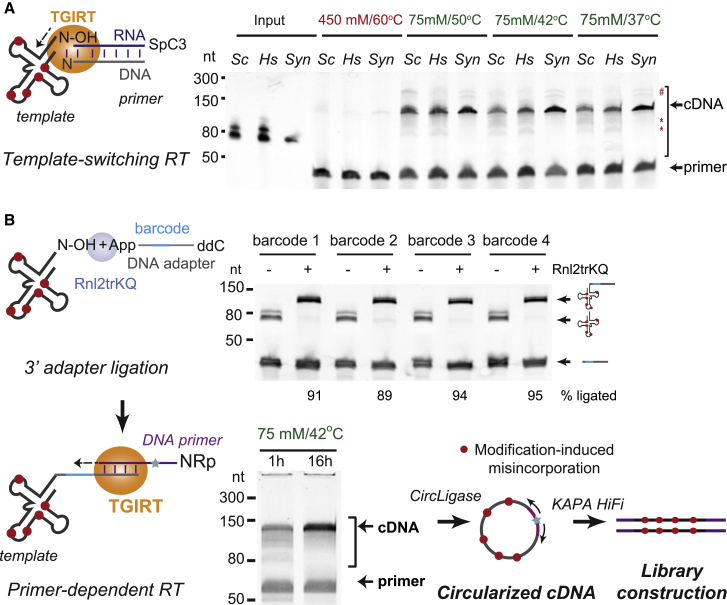
(A) Schematic of template-switching TGIRT reactions primed by an RNA/DNA duplex with a single-nucleotide 3′ overhang and a gel image of cDNA products from endogenously modified tRNA pools from S. cerevisiae (Sc), K562 cells (Hs), or a synthetic unmodified tRNA (Syn) at different reaction temperatures and salt concentration. Red, reaction conditions previously used for tRNA library construction; asterisks, premature stops to cDNA synthesis; hash, potential products from end-to-end linkage of tRNAs.
(B) Schematic of the mim-tRNAseq library generation workflow. Top gel image: 3′ adapter ligation reactions with four barcoded adapters. Ligation efficiency was measured by normalizing input tRNA band intensity to that in reactions from which Rnl2trKQ was omitted. Bottom gel image: comparison of cDNA yield in short (1 h) or extended (16 h) primer-dependent TGIRT RT on a mix of adapter-ligated tRNA pools from S. cerevisiae and human K562 and HEK293T cells.
See also Figure S1 and STAR methods.